NPs Basic Information
|
Name |
Actinoallolide B
|
| Molecular Formula | C32H54O8 | |
| IUPAC Name* |
(5R,12R)-12-ethyl-1-hydroxy-8,10-dimethyl-5-[(2R,3R,4R,8S,9R,10R,11S)-3,9,11-trihydroxy-4,6,8,10-tetramethyltridec-6-en-2-yl]-4,13-dioxabicyclo[8.2.1]tridec-7-ene-3,11-dione
|
|
| SMILES |
CC[C@@H]1C(=O)C2(CC(=CC[C@@H](OC(=O)CC1(O2)O)[C@H](C)[C@@H]([C@H](C)CC(=C[C@H](C)[C@H]([C@H](C)[C@H](CC)O)O)C)O)C)C
|
|
| InChI |
InChI=1S/C32H54O8/c1-10-24-30(37)31(9)16-18(3)12-13-26(39-27(34)17-32(24,38)40-31)23(8)29(36)21(6)15-19(4)14-20(5)28(35)22(7)25(33)11-2/h12,14,20-26,28-29,33,35-36,38H,10-11,13,15-17H2,1-9H3/t20-,21+,22+,23-,24+,25-,26+,28+,29+,31?,32?/m0/s1
|
|
| InChIKey |
GLIPEBQBWMVZJH-CPMPDNOVSA-N
|
|
| Synonyms |
Actinoallolide B
|
|
| CAS | NA | |
| PubChem CID | 156580534 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 566.8 | ALogp: | 4.6 |
| HBD: | 4 | HBA: | 8 |
| Rotatable Bonds: | 11 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 134.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 40 | QED Weighted: | 0.203 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.727 | MDCK Permeability: | 0.00002750 |
| Pgp-inhibitor: | 0.997 | Pgp-substrate: | 0.986 |
| Human Intestinal Absorption (HIA): | 0.298 | 20% Bioavailability (F20%): | 0.093 |
| 30% Bioavailability (F30%): | 0.324 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.003 | Plasma Protein Binding (PPB): | 89.32% |
| Volume Distribution (VD): | 0.974 | Fu: | 5.51% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.013 | CYP1A2-substrate: | 0.095 |
| CYP2C19-inhibitor: | 0.009 | CYP2C19-substrate: | 0.882 |
| CYP2C9-inhibitor: | 0.008 | CYP2C9-substrate: | 0.073 |
| CYP2D6-inhibitor: | 0.001 | CYP2D6-substrate: | 0.05 |
| CYP3A4-inhibitor: | 0.454 | CYP3A4-substrate: | 0.649 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.287 | Half-life (T1/2): | 0.57 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.011 | Human Hepatotoxicity (H-HT): | 0.929 |
| Drug-inuced Liver Injury (DILI): | 0.879 | AMES Toxicity: | 0.063 |
| Rat Oral Acute Toxicity: | 0.914 | Maximum Recommended Daily Dose: | 0.02 |
| Skin Sensitization: | 0.056 | Carcinogencity: | 0.068 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.009 |
| Respiratory Toxicity: | 0.12 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
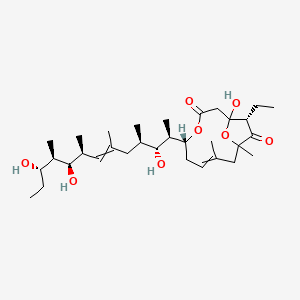
| ENC004257 | 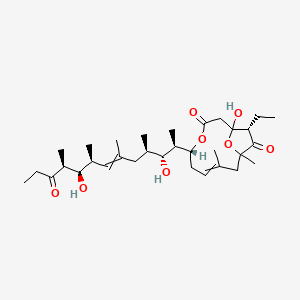 |
0.826 | D0Y7LD |  |
0.190 | ||
| ENC004259 |  |
0.719 | D06LNW |  |
0.188 | ||
| ENC004260 |  |
0.588 | D0W2EK | 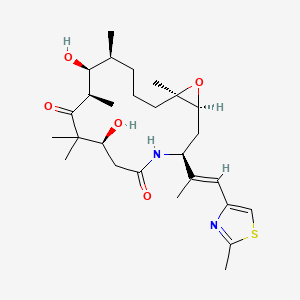 |
0.185 | ||
| ENC004255 |  |
0.434 | D0G6AB | 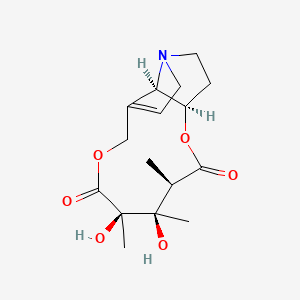 |
0.184 | ||
| ENC003822 |  |
0.281 | D0E9KA | 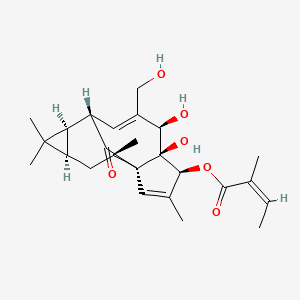 |
0.184 | ||
| ENC003155 |  |
0.275 | D0H2MO | 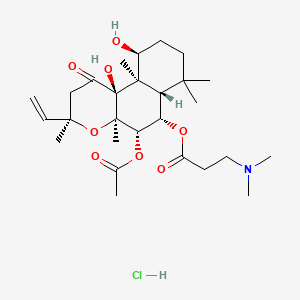 |
0.183 | ||
| ENC003821 |  |
0.273 | D0WY9N |  |
0.177 | ||
| ENC005126 | 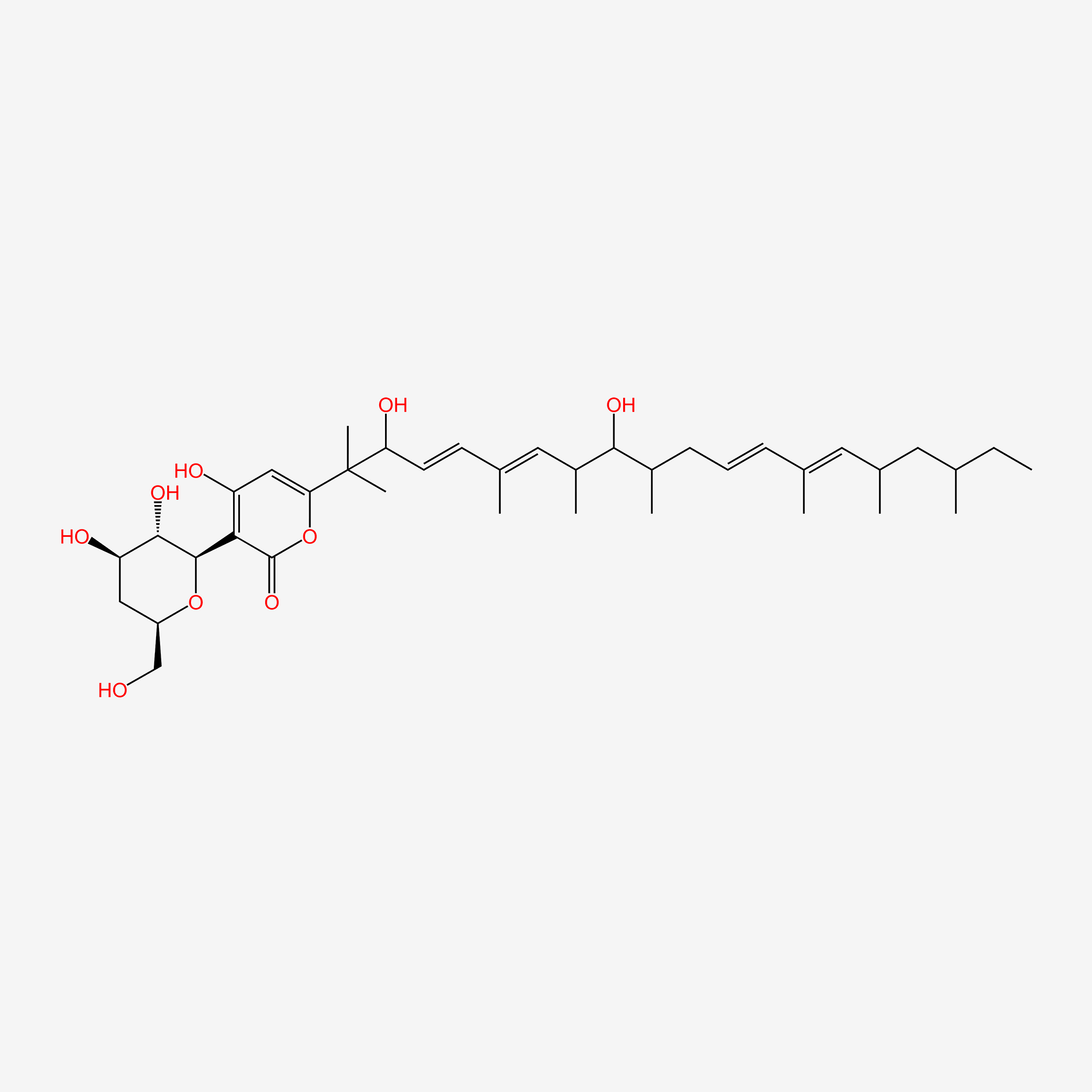 |
0.271 | D08SVH | 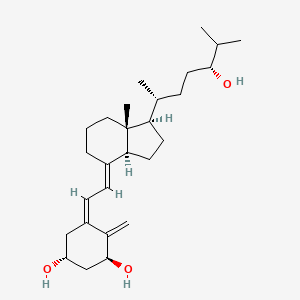 |
0.176 | ||
| ENC002304 |  |
0.253 | D03KIA |  |
0.176 | ||
| ENC004454 |  |
0.246 | D0X1WJ |  |
0.176 | ||