NPs Basic Information

|
Name |
Methyl 3,4-dihydroxyphenylacetate
|
| Molecular Formula | C9H10O4 | |
| IUPAC Name* |
methyl 2-(3,4-dihydroxyphenyl)acetate
|
|
| SMILES |
COC(=O)CC1=CC(=C(C=C1)O)O
|
|
| InChI |
InChI=1S/C9H10O4/c1-13-9(12)5-6-2-3-7(10)8(11)4-6/h2-4,10-11H,5H2,1H3
|
|
| InChIKey |
UGFILLIGHGZLHE-UHFFFAOYSA-N
|
|
| Synonyms |
25379-88-8; Methyl 2-(3,4-dihydroxyphenyl)acetate; Methyl 3,4-dihydroxyphenylacetate; 3,4-Dihydroxyphenylacetic Acid Methyl Ester; CHEMBL3120494; Benzeneacetic acid, 3,4-dihydroxy-, methyl ester; Homoprotocatechuic Acid Methyl Ester; methyl (3,4-dihydroxyphenyl)acetate; methyl homoprotocatechuate; SCHEMBL3668882; DTXSID70451666; ZINC2566703; BDBM50062147; Catechol-4-acetic Acid Methyl Ester; MFCD00583539; AKOS015900183; Methyl2-(3,4-dihydroxyphenyl)acetate; CS-W017123; Pyrocatechol-4-acetic Acid Methyl Ester; AS-44408; ERGOLINE-8-CARBOXYLICACIDMETHYLESTER; D2734; FT-0638516; (3,4-dihydroxyphenyl)-acetic acid methyl ester; EN300-732967; 379D888; A817833; J-015971
|
|
| CAS | 25379-88-8 | |
| PubChem CID | 11008556 | |
| ChEMBL ID | CHEMBL3120494 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 182.17 | ALogp: | 0.8 |
| HBD: | 2 | HBA: | 4 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 66.8 | Aromatic Rings: | 1 |
| Heavy Atoms: | 13 | QED Weighted: | 0.533 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.551 | MDCK Permeability: | 0.00002090 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.059 |
| Human Intestinal Absorption (HIA): | 0.018 | 20% Bioavailability (F20%): | 0.005 |
| 30% Bioavailability (F30%): | 0.786 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.095 | Plasma Protein Binding (PPB): | 45.83% |
| Volume Distribution (VD): | 0.617 | Fu: | 50.12% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.323 | CYP1A2-substrate: | 0.523 |
| CYP2C19-inhibitor: | 0.166 | CYP2C19-substrate: | 0.098 |
| CYP2C9-inhibitor: | 0.211 | CYP2C9-substrate: | 0.805 |
| CYP2D6-inhibitor: | 0.068 | CYP2D6-substrate: | 0.504 |
| CYP3A4-inhibitor: | 0.182 | CYP3A4-substrate: | 0.257 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 17.862 | Half-life (T1/2): | 0.932 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.022 | Human Hepatotoxicity (H-HT): | 0.128 |
| Drug-inuced Liver Injury (DILI): | 0.563 | AMES Toxicity: | 0.488 |
| Rat Oral Acute Toxicity: | 0.1 | Maximum Recommended Daily Dose: | 0.014 |
| Skin Sensitization: | 0.612 | Carcinogencity: | 0.176 |
| Eye Corrosion: | 0.503 | Eye Irritation: | 0.928 |
| Respiratory Toxicity: | 0.184 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000035 |  |
0.675 | D0U0OT | 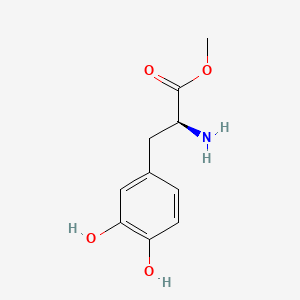 |
0.630 | ||
| ENC004860 |  |
0.545 | D0BA6T | 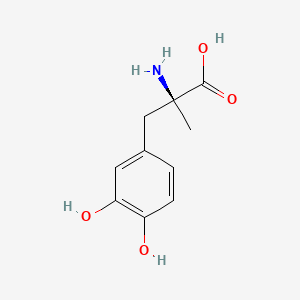 |
0.542 | ||
| ENC000127 |  |
0.532 | D08HVR | 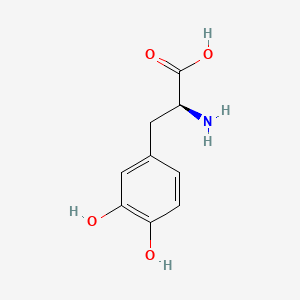 |
0.532 | ||
| ENC000862 |  |
0.489 | D0T7OW |  |
0.512 | ||
| ENC000002 |  |
0.488 | D0P7JZ | 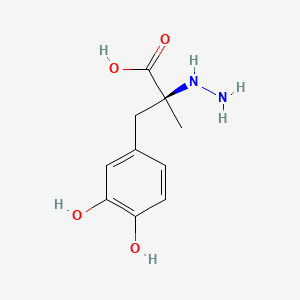 |
0.510 | ||
| ENC002242 |  |
0.479 | D0Y6KO | 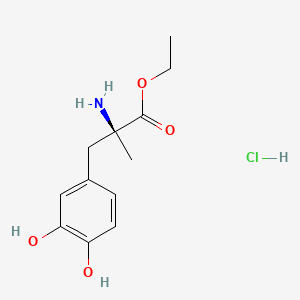 |
0.500 | ||
| ENC000329 |  |
0.475 | D0V9EN | 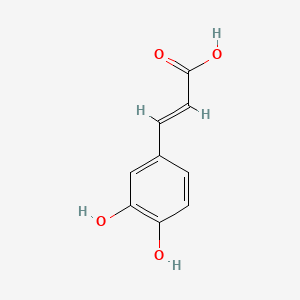 |
0.429 | ||
| ENC002317 |  |
0.457 | D04PHC | 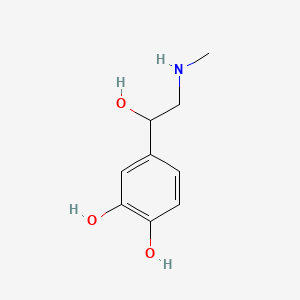 |
0.400 | ||
| ENC004024 |  |
0.446 | D0I3RO | 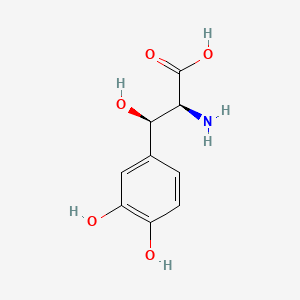 |
0.396 | ||
| ENC000325 |  |
0.442 | D07MOX |  |
0.396 | ||