NPs Basic Information

|
Name |
Azulene
|
| Molecular Formula | C10H8 | |
| IUPAC Name* |
azulene
|
|
| SMILES |
C1=CC=C2C=CC=C2C=C1
|
|
| InChI |
InChI=1S/C10H8/c1-2-5-9-7-4-8-10(9)6-3-1/h1-8H
|
|
| InChIKey |
CUFNKYGDVFVPHO-UHFFFAOYSA-N
|
|
| Synonyms |
azulene; 275-51-4; Cyclopentacycloheptene; Azunamic; Bicyclo[5.3.0]decapentaene; azulen; Bicyclo(5.3.0)decapentaene; Bicyclo(5.3.0)-1,3,5,7,9-decapentaene; Bicyclo(5.3.0)-deca-2,4,6,8,10-pentaene; 82R6M9MGLP; CHEBI:31249; Azulene (JAN); NSC-89248; AZULENE [JAN]; EINECS 205-993-6; 82451-56-7; MFCD00003810; NSC 89248; UNII-82R6M9MGLP; Azulene, 99%; AZULENE [INCI]; AZULENE [MI]; AZULENE [MART.]; AZULENE [WHO-DD]; bicyclo(5.3.0)-deca-1,3,5,7,9-pentaene; Azulene, analytical standard; Azusalen [as sodium sulfonate]; CHEMBL3272628; DTXSID2059770; HY-B0055; NSC89248; ZINC1570209; AKOS015840881; CS-15638; Bicyclo[5.3.0]-1,5,7,9-decapentaene; DB-047243; A0634; Bicyclo(0.3.5)deca-1,3,5,7,9-pentaene; CS-0006517; FT-0622537; Azulene, standard for GC, >=99.0% (GC); bicyclo[5.3.0]deca-2,4,6,8,10-pentaene; C13408; D09768; A819116; Q144362; SR-01000944574; J-016811; SR-01000944574-1; BICYCLO-(0.3.5)-DECA-1,3,5,7,9-PENTAENE; BICYCLO-(5.3.0)-DECA-2,4,6,8,10-PENTAENE
|
|
| CAS | 275-51-4 | |
| PubChem CID | 9231 | |
| ChEMBL ID | CHEMBL3272628 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 128.17 | ALogp: | 3.2 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 10 | QED Weighted: | 0.508 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.238 | MDCK Permeability: | 0.00002060 |
| Pgp-inhibitor: | 0.003 | Pgp-substrate: | 0.046 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.988 |
| 30% Bioavailability (F30%): | 0.216 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.469 | Plasma Protein Binding (PPB): | 95.14% |
| Volume Distribution (VD): | 1.172 | Fu: | 5.02% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.993 | CYP1A2-substrate: | 0.512 |
| CYP2C19-inhibitor: | 0.715 | CYP2C19-substrate: | 0.082 |
| CYP2C9-inhibitor: | 0.345 | CYP2C9-substrate: | 0.431 |
| CYP2D6-inhibitor: | 0.02 | CYP2D6-substrate: | 0.31 |
| CYP3A4-inhibitor: | 0.015 | CYP3A4-substrate: | 0.241 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.714 | Half-life (T1/2): | 0.385 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.077 | Human Hepatotoxicity (H-HT): | 0.049 |
| Drug-inuced Liver Injury (DILI): | 0.232 | AMES Toxicity: | 0.726 |
| Rat Oral Acute Toxicity: | 0.215 | Maximum Recommended Daily Dose: | 0.05 |
| Skin Sensitization: | 0.629 | Carcinogencity: | 0.763 |
| Eye Corrosion: | 0.842 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.093 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000047 |  |
0.500 | D0Y7EM | 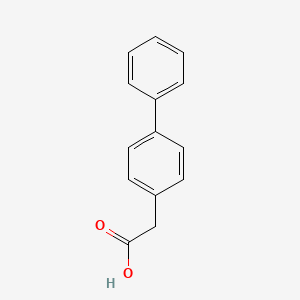 |
0.520 | ||
| ENC001050 | 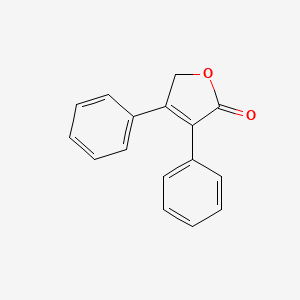 |
0.482 | D06LHG |  |
0.448 | ||
| ENC001388 | 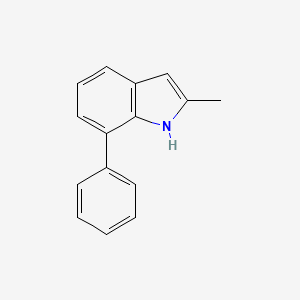 |
0.481 | D0A1PX | 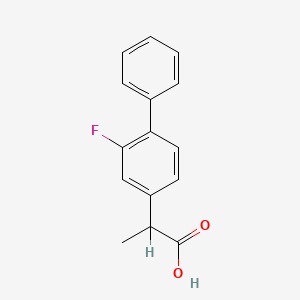 |
0.404 | ||
| ENC000732 |  |
0.453 | D0M9DC | 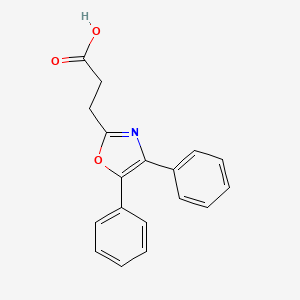 |
0.403 | ||
| ENC000167 | 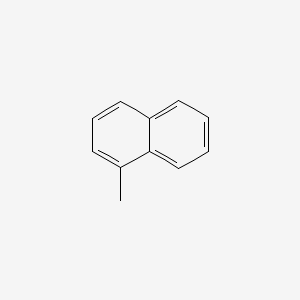 |
0.442 | D02WCI |  |
0.400 | ||
| ENC000093 | 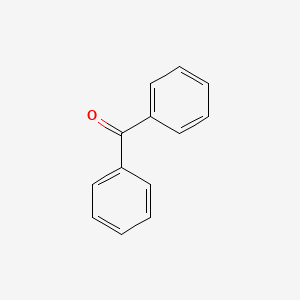 |
0.420 | D0B1FE | 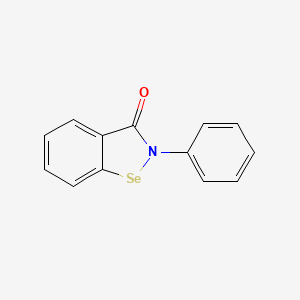 |
0.375 | ||
| ENC002076 | 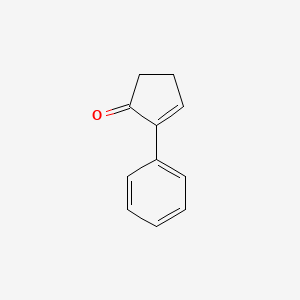 |
0.413 | D0G1VX | 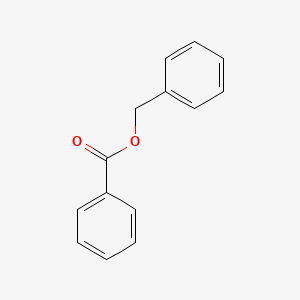 |
0.375 | ||
| ENC000461 | 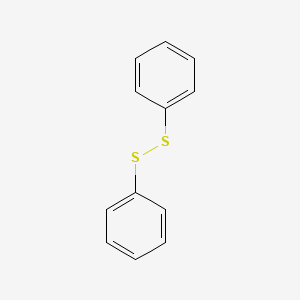 |
0.412 | D03XYW | 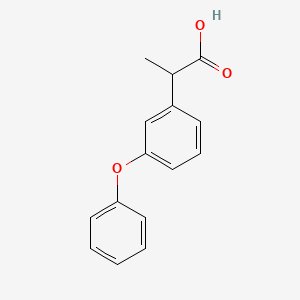 |
0.373 | ||
| ENC000159 |  |
0.408 | D0H6TP |  |
0.370 | ||
| ENC000036 |  |
0.408 | D08FTG |  |
0.362 | ||