NPs Basic Information

|
Name |
1-Methyl-2-phenylindole
|
| Molecular Formula | C15H13N | |
| IUPAC Name* |
1-methyl-2-phenylindole
|
|
| SMILES |
CN1C2=CC=CC=C2C=C1C3=CC=CC=C3
|
|
| InChI |
InChI=1S/C15H13N/c1-16-14-10-6-5-9-13(14)11-15(16)12-7-3-2-4-8-12/h2-11H,1H3
|
|
| InChIKey |
SFWZZSXCWQTORH-UHFFFAOYSA-N
|
|
| Synonyms |
1-Methyl-2-phenylindole; 3558-24-5; 1-Methyl-2-phenyl-1H-indole; 1H-Indole, 1-methyl-2-phenyl-; 2-Phenyl-N-methylindole; N-Methyl-2-phenylindole; Indole, 1-methyl-2-phenyl-; IE3UB978IA; MFCD00022892; NSC-63793; NSC63793; EINECS 222-618-1; NSC 63793; 2-phenyl-1-methylindole; n-methyl-2-phenyl indole; n-methyl-2-phenyl-indole; 1-methyl-2-phenyl-indole; UNII-IE3UB978IA; NCIOpen2_002836; MLS001215074; 1-methyl-2-phenyl-1H-indol; BIDD:GT0516; SCHEMBL214075; CHEMBL1545735; DTXSID5063073; 1-Methyl-2-phenylindole, 99%; 1-Methyl-2-phenyl-1H-indole #; HMS2857M05; AMY23168; ZINC1081188; AC8408; BBL012866; STK202660; AKOS001426229; CS-W015261; NCGC00246339-01; AC-10577; AS-59068; BP-13058; SMR000543310; SY038925; DB-019242; FT-0608050; M0733; EN300-20675; AB00775587-01; A822887; AF-963/00522046; J-504920; Z104479722
|
|
| CAS | 3558-24-5 | |
| PubChem CID | 77095 | |
| ChEMBL ID | CHEMBL1545735 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 207.27 | ALogp: | 4.1 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 4.9 | Aromatic Rings: | 3 |
| Heavy Atoms: | 16 | QED Weighted: | 0.553 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.621 | MDCK Permeability: | 0.00002270 |
| Pgp-inhibitor: | 0.014 | Pgp-substrate: | 0.128 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.912 |
| 30% Bioavailability (F30%): | 0.006 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.843 | Plasma Protein Binding (PPB): | 96.96% |
| Volume Distribution (VD): | 1.153 | Fu: | 2.10% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.991 | CYP1A2-substrate: | 0.643 |
| CYP2C19-inhibitor: | 0.846 | CYP2C19-substrate: | 0.161 |
| CYP2C9-inhibitor: | 0.503 | CYP2C9-substrate: | 0.609 |
| CYP2D6-inhibitor: | 0.132 | CYP2D6-substrate: | 0.776 |
| CYP3A4-inhibitor: | 0.048 | CYP3A4-substrate: | 0.249 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.657 | Half-life (T1/2): | 0.22 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.049 | Human Hepatotoxicity (H-HT): | 0.09 |
| Drug-inuced Liver Injury (DILI): | 0.172 | AMES Toxicity: | 0.878 |
| Rat Oral Acute Toxicity: | 0.226 | Maximum Recommended Daily Dose: | 0.712 |
| Skin Sensitization: | 0.429 | Carcinogencity: | 0.838 |
| Eye Corrosion: | 0.041 | Eye Irritation: | 0.976 |
| Respiratory Toxicity: | 0.13 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001388 | 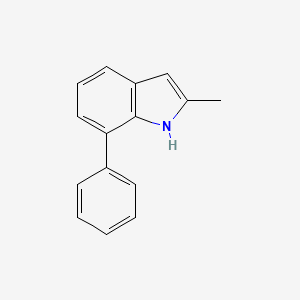 |
0.492 | D0B1FE | 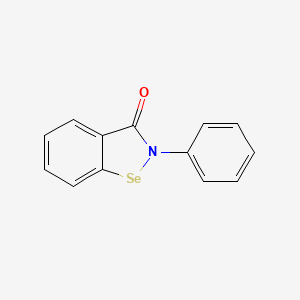 |
0.424 | ||
| ENC000321 |  |
0.453 | D09VXM | 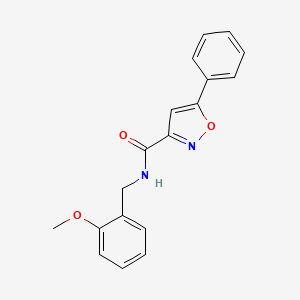 |
0.407 | ||
| ENC000047 |  |
0.453 | D06UDO | 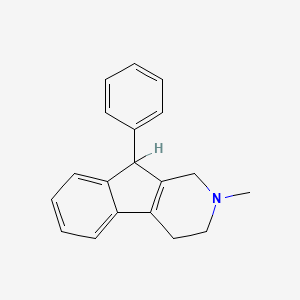 |
0.395 | ||
| ENC001050 | 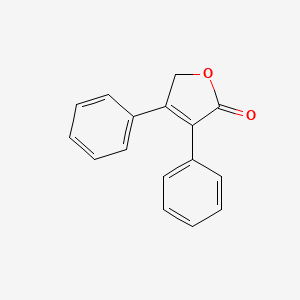 |
0.449 | D05AFX | 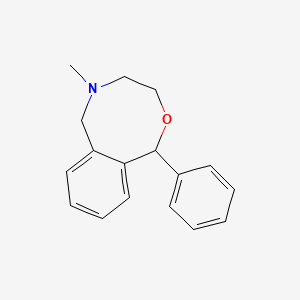 |
0.392 | ||
| ENC001109 |  |
0.437 | D08FTG | 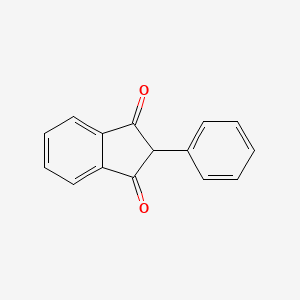 |
0.391 | ||
| ENC000093 | 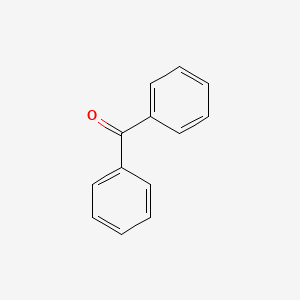 |
0.419 | D0M9DC | 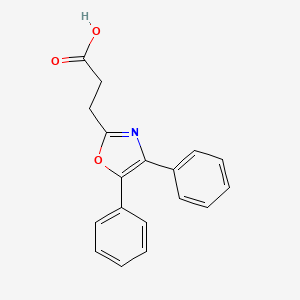 |
0.388 | ||
| ENC003032 | 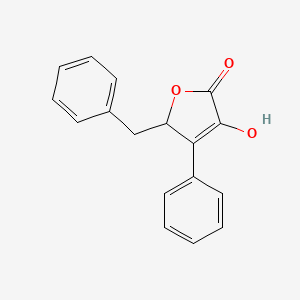 |
0.419 | D07JVL | 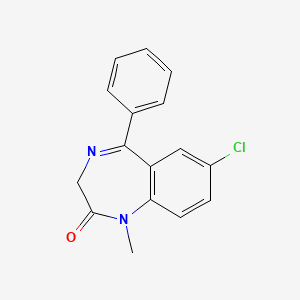 |
0.387 | ||
| ENC004519 | 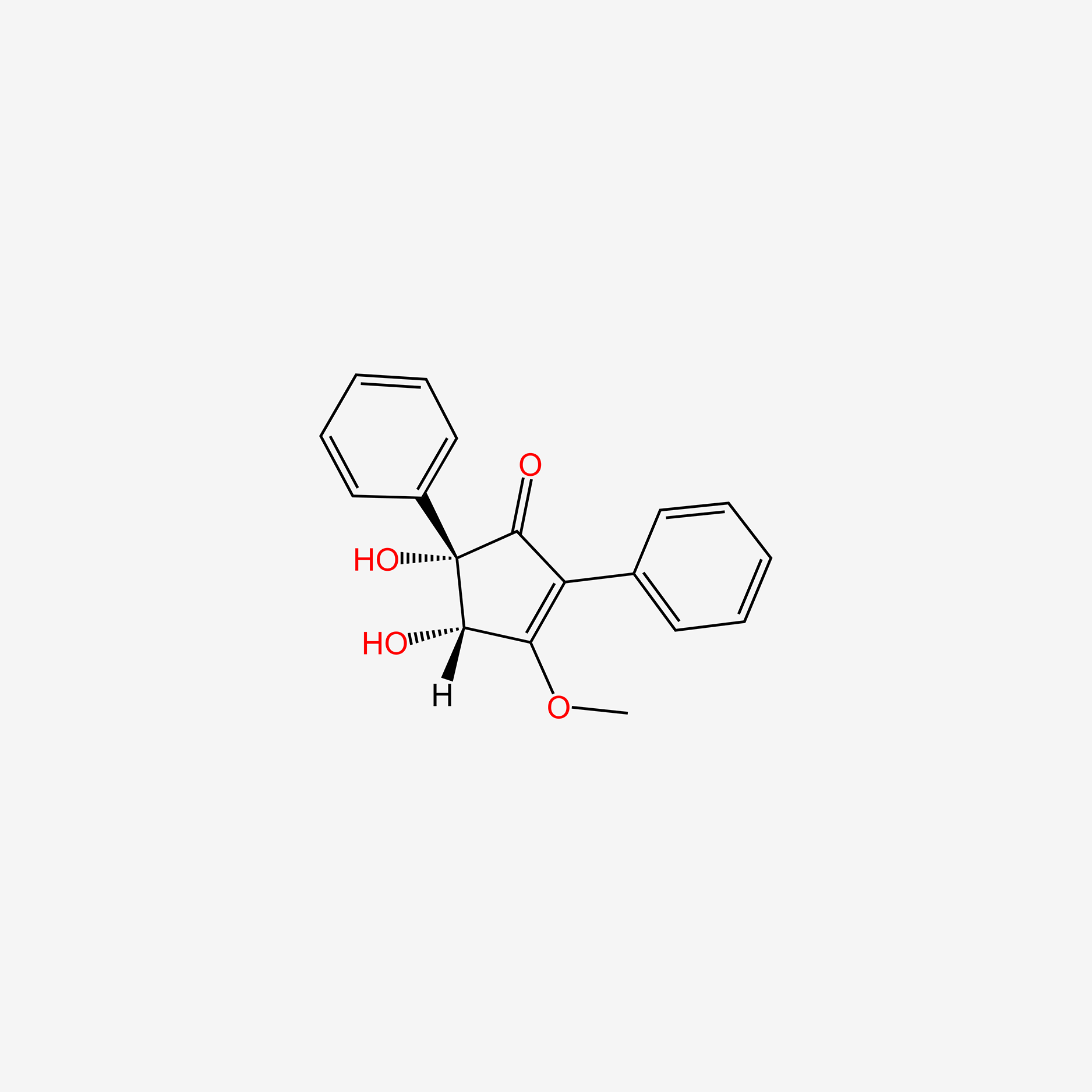 |
0.416 | D0A1PX | 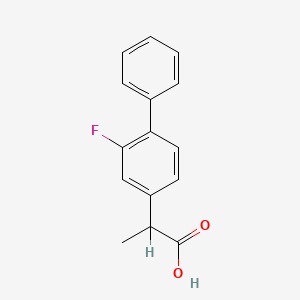 |
0.386 | ||
| ENC004518 | 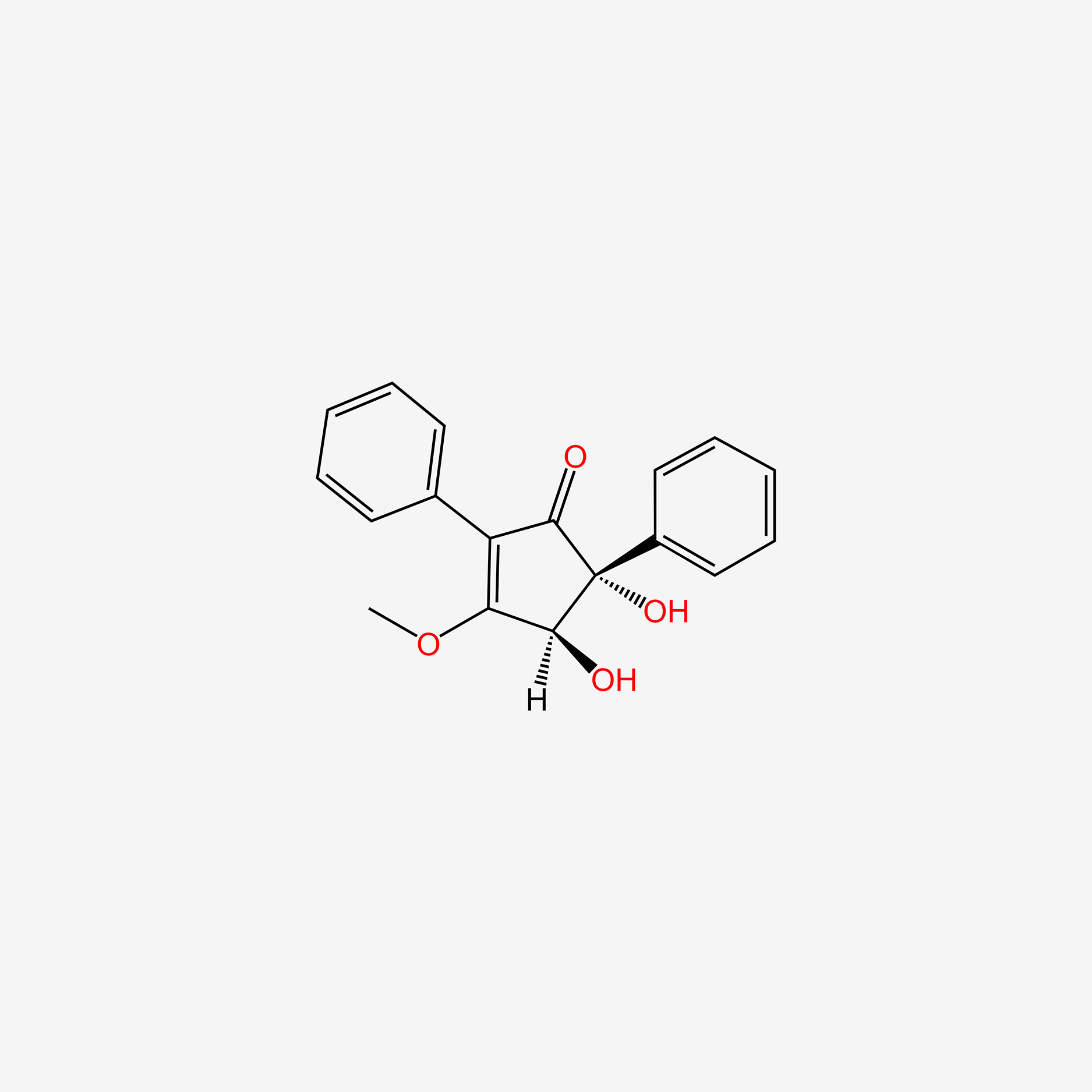 |
0.416 | D00HPK | 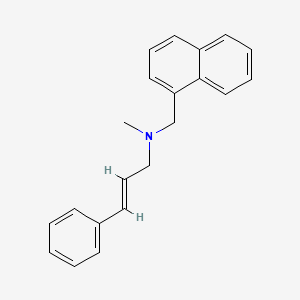 |
0.383 | ||
| ENC004517 | 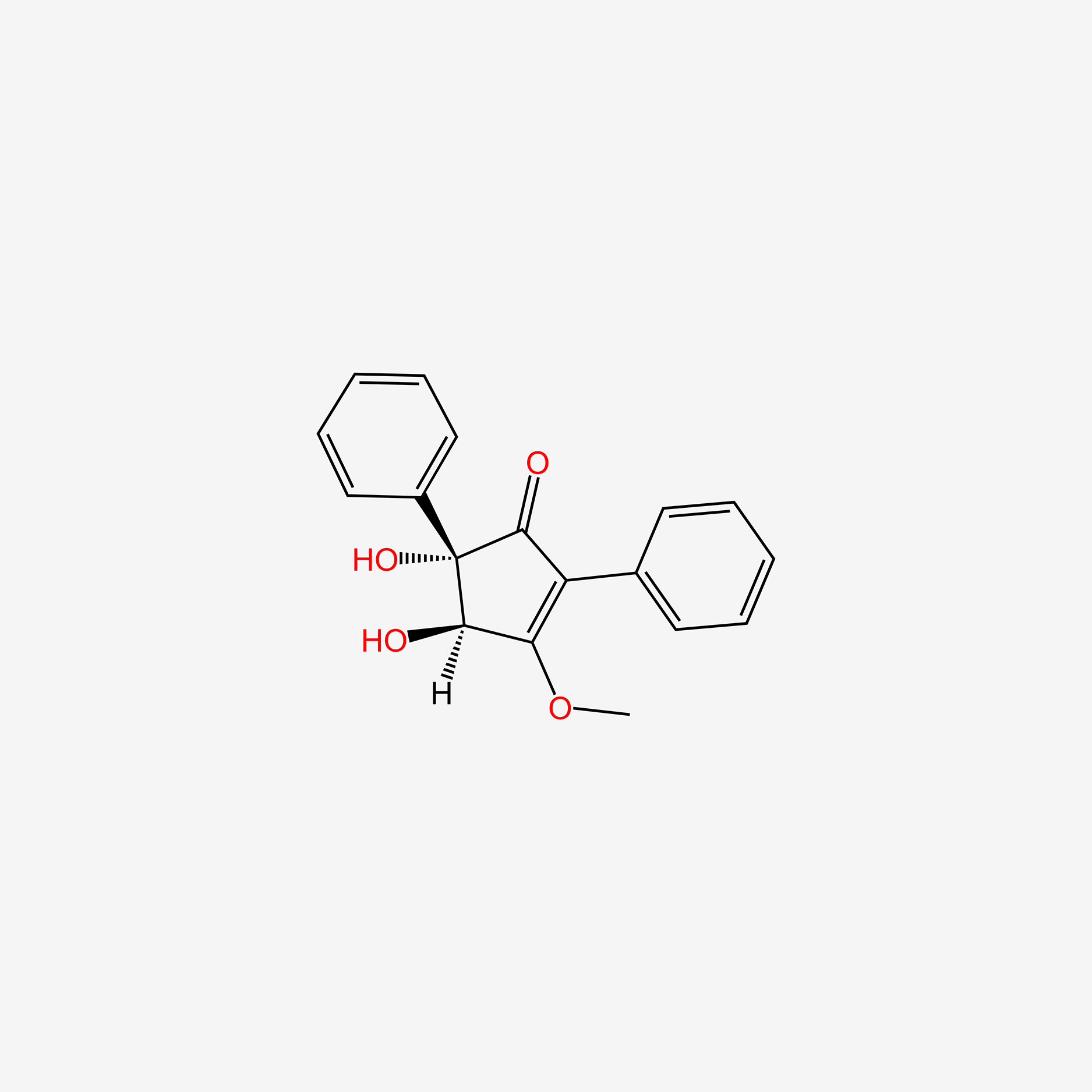 |
0.416 | D0G1VX | 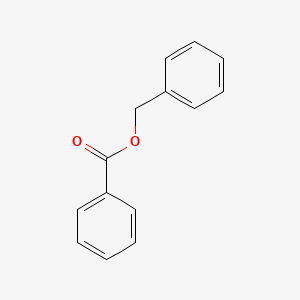 |
0.382 | ||