NPs Basic Information

|
Name |
beta-Ocimene
|
| Molecular Formula | C10H16 | |
| IUPAC Name* |
(3E)-3,7-dimethylocta-1,3,6-triene
|
|
| SMILES |
CC(=CC/C=C(\C)/C=C)C
|
|
| InChI |
InChI=1S/C10H16/c1-5-10(4)8-6-7-9(2)3/h5,7-8H,1,6H2,2-4H3/b10-8+
|
|
| InChIKey |
IHPKGUQCSIINRJ-CSKARUKUSA-N
|
|
| Synonyms |
(E)-beta-ocimene; OCIMENE; trans-beta-Ocimene; 3779-61-1; (E)-3,7-Dimethylocta-1,3,6-triene; (3E)-3,7-dimethylocta-1,3,6-triene; 1,3,6-Octatriene, 3,7-dimethyl-; beta-Ocimene; 13877-91-3; trans-Ocimene; Ocimene, beta-; 3,7-DIMETHYL-1,3,6-OCTATRIENE; Ocimene trans-beta-form; 3,7-dimethyl-1,3E,6-octatriene; .beta.-Ocimene; 1,3,6-Octatriene, 3,7-dimethyl-, (E)-; (E)-Ocimene; FEMA No. 3539; .beta.-trans-Ocimene; trans-.beta.-Ocimene; Ocimene, trans-.beta.-; trans-3,7-Dimethyl-1,3,6-Octatriene; (3E)-3,7-Dimethyl-1,3,6-octatriene; (E)-3,7-Dimethyloctatriene; 38BQ4UYY06; 3,7-Dimethyl-1,3,6-octatriene (natural); CHEBI:64280; 3,7-DIMETHYLOCTA-1,3,6-TRIENE, (E)-; E-3,7-Dimethyl-1,3,6-octatriene; trans-3,7-dimethylocta-1,3,6-triene; UNII-38BQ4UYY06; UNII-V96I2PX4L5; 1,3,6-Octatriene,3,7-dimethyl-; p-Ocimene; Ocimene, trans; E-beta-Ocimene; beta-trans-Ocimene; t-.beta.-Ocimene; Ocimene (E); EINECS 223-241-5; EINECS 237-641-2; beta -trans-Ocimene; trans-beta -Ocimene; ocimene (e-beta-); (3E)-beta-ocimene; (E)-beta -Ocimene; beta -(E)-Ocimene; .beta.-(E)-Ocimene; (E)- .beta.-Ocimene; .beta.-Ocimene, (E)-; bmse000964; 1,3,6-Octatriene, 3,7-dimethyl-, (3E)-; 3,7-Dimethyl-(E)-Octatriene; V96I2PX4L5; CHEMBL2228374; DTXSID8040567; OCIMENE TRANS-.BETA.-FORM; .BETA.-OCIMENE, (3E)-; ZINC1531618; Octatriene, 3,7-dimethyl-, (E)-; AKOS015903787; (e)-3,7-dimethyl-1,3,6-octatriene; LMPR0102010021; 3,7-Dimethyl-(E)-1,3,6-Octatriene; beta-Ocimene 100 microg/mL in Methanol; OCIMENE TRANS-.BETA.-FORM [MI]; AS-80273; (3E)-3,7-dimethyl-octa-1,3,6-triene; DIMETHYL-1,3,6-OCTATRIENE [FHFI]; C09873; 3,7-DIMETHYLOCTA-1,3,6-TRIENE, (3E)-; J-007176; Q26777012
|
|
| CAS | 3779-61-1 | |
| PubChem CID | 5281553 | |
| ChEMBL ID | CHEMBL2228374 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 136.23 | ALogp: | 4.3 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 3 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 10 | QED Weighted: | 0.402 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.427 | MDCK Permeability: | 0.00003080 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.091 |
| 30% Bioavailability (F30%): | 0.436 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.943 | Plasma Protein Binding (PPB): | 93.30% |
| Volume Distribution (VD): | 3.872 | Fu: | 8.52% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.777 | CYP1A2-substrate: | 0.581 |
| CYP2C19-inhibitor: | 0.31 | CYP2C19-substrate: | 0.854 |
| CYP2C9-inhibitor: | 0.082 | CYP2C9-substrate: | 0.906 |
| CYP2D6-inhibitor: | 0.079 | CYP2D6-substrate: | 0.787 |
| CYP3A4-inhibitor: | 0.047 | CYP3A4-substrate: | 0.259 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.401 | Half-life (T1/2): | 0.684 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.827 |
| Drug-inuced Liver Injury (DILI): | 0.042 | AMES Toxicity: | 0.19 |
| Rat Oral Acute Toxicity: | 0.04 | Maximum Recommended Daily Dose: | 0.213 |
| Skin Sensitization: | 0.892 | Carcinogencity: | 0.845 |
| Eye Corrosion: | 0.976 | Eye Irritation: | 0.992 |
| Respiratory Toxicity: | 0.925 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000526 | 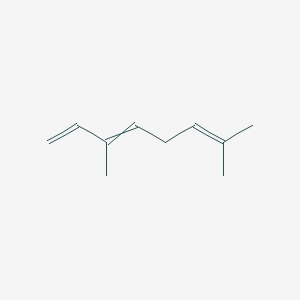 |
1.000 | D0M1PQ |  |
0.268 | ||
| ENC001564 | 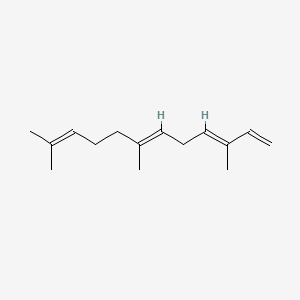 |
0.610 | D0H6VY | 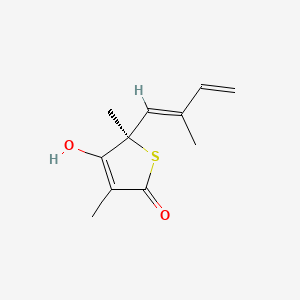 |
0.240 | ||
| ENC001664 | 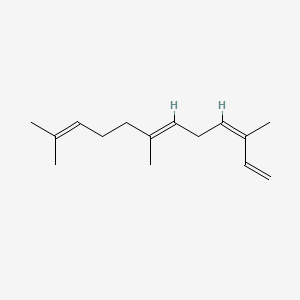 |
0.610 | D05XQE |  |
0.229 | ||
| ENC001566 | 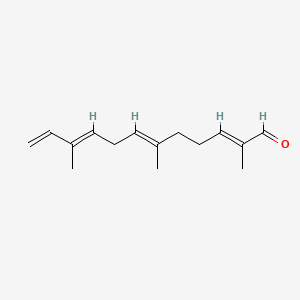 |
0.500 | D09XWD |  |
0.213 | ||
| ENC002306 | 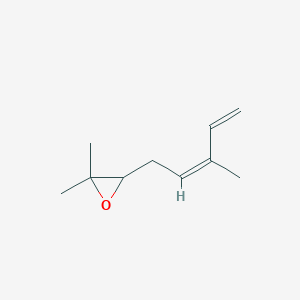 |
0.385 | D0S7WX | 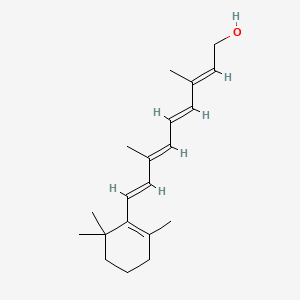 |
0.186 | ||
| ENC001434 | 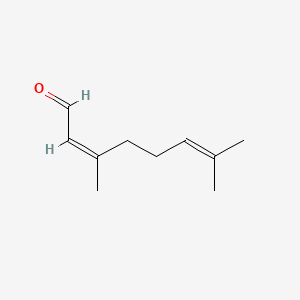 |
0.375 | D03VFL | 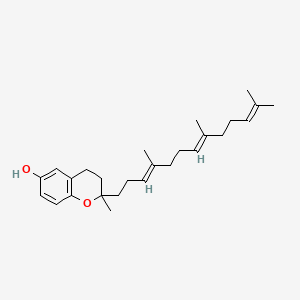 |
0.182 | ||
| ENC001424 | 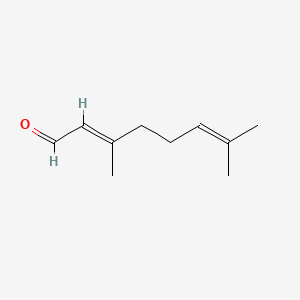 |
0.375 | D0F1GS |  |
0.162 | ||
| ENC001718 | 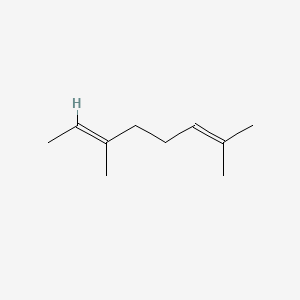 |
0.368 | D0Z4NI |  |
0.162 | ||
| ENC001641 | 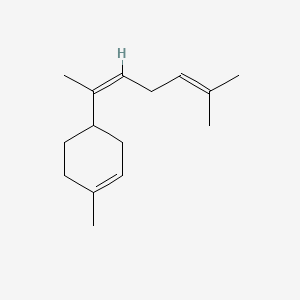 |
0.367 | D0Q6DX |  |
0.161 | ||
| ENC000314 | 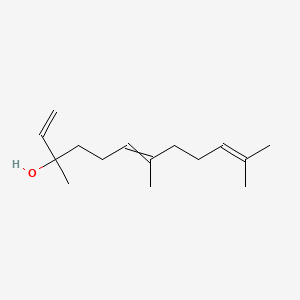 |
0.360 | D06BLQ |  |
0.157 | ||