NPs Basic Information
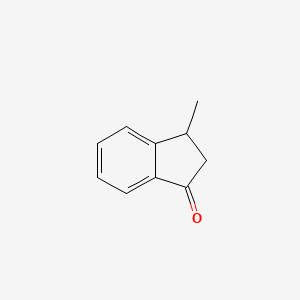
|
Name |
3-Methyl-1-indanone
|
| Molecular Formula | C10H10O | |
| IUPAC Name* |
3-methyl-2,3-dihydroinden-1-one
|
|
| SMILES |
CC1CC(=O)C2=CC=CC=C12
|
|
| InChI |
InChI=1S/C10H10O/c1-7-6-10(11)9-5-3-2-4-8(7)9/h2-5,7H,6H2,1H3
|
|
| InChIKey |
XVTQSYKCADSUHN-UHFFFAOYSA-N
|
|
| Synonyms |
6072-57-7; 3-Methyl-1-indanone; 3-Methyl-2,3-dihydro-1H-inden-1-one; 3-Methylindan-1-One; 3-Methylindanone; 3-methyl-2,3-dihydroinden-1-one; 3-Methyl-indan-1-one; 1H-Inden-1-one, 2,3-dihydro-3-methyl-; 1-Indanone, 3-methyl-; 2,3-Dihydro-3-methyl-1H-inden-1-one; XJW7YA5CLT; NSC-89554; NSC89554; UNII-XJW7YA5CLT; SCHEMBL181579; 3-Methyl-1-indanone, 99%; SCHEMBL20368206; 3-methyl-2,3-dihydro-inden-1-one; AM1094; MFCD00156725; NSC 89554; 3-METHYLINDANONE, (+/-)-; AKOS000249461; AKOS016038081; 1H-Inden-1-one,3-dihydro-3-methyl-; AS-5626; AC-23412; DB-001259; FT-0659583; EN300-66017; H10914; 072M577; F8884-2040; Z335243948
|
|
| CAS | 6072-57-7 | |
| PubChem CID | 259560 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 146.19 | ALogp: | 2.0 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 2 |
| Heavy Atoms: | 11 | QED Weighted: | 0.549 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.481 | MDCK Permeability: | 0.00002520 |
| Pgp-inhibitor: | 0.01 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.008 |
| 30% Bioavailability (F30%): | 0.023 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.933 | Plasma Protein Binding (PPB): | 75.62% |
| Volume Distribution (VD): | 0.649 | Fu: | 23.58% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.904 | CYP1A2-substrate: | 0.931 |
| CYP2C19-inhibitor: | 0.791 | CYP2C19-substrate: | 0.498 |
| CYP2C9-inhibitor: | 0.305 | CYP2C9-substrate: | 0.483 |
| CYP2D6-inhibitor: | 0.047 | CYP2D6-substrate: | 0.683 |
| CYP3A4-inhibitor: | 0.079 | CYP3A4-substrate: | 0.274 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.149 | Half-life (T1/2): | 0.328 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.014 | Human Hepatotoxicity (H-HT): | 0.098 |
| Drug-inuced Liver Injury (DILI): | 0.594 | AMES Toxicity: | 0.809 |
| Rat Oral Acute Toxicity: | 0.272 | Maximum Recommended Daily Dose: | 0.564 |
| Skin Sensitization: | 0.185 | Carcinogencity: | 0.847 |
| Eye Corrosion: | 0.099 | Eye Irritation: | 0.977 |
| Respiratory Toxicity: | 0.71 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC004792 | 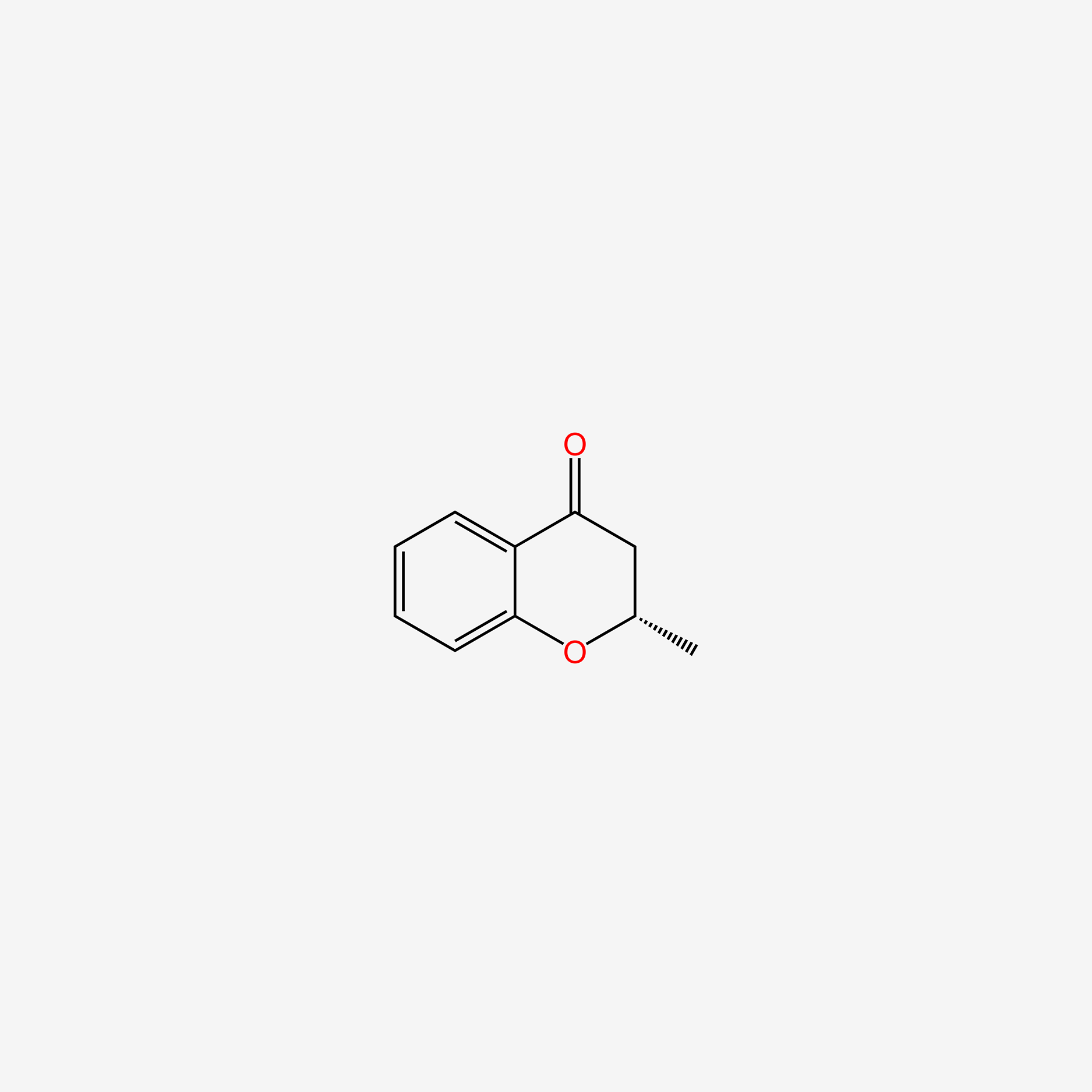 |
0.585 | D06BYV |  |
0.373 | ||
| ENC006142 | 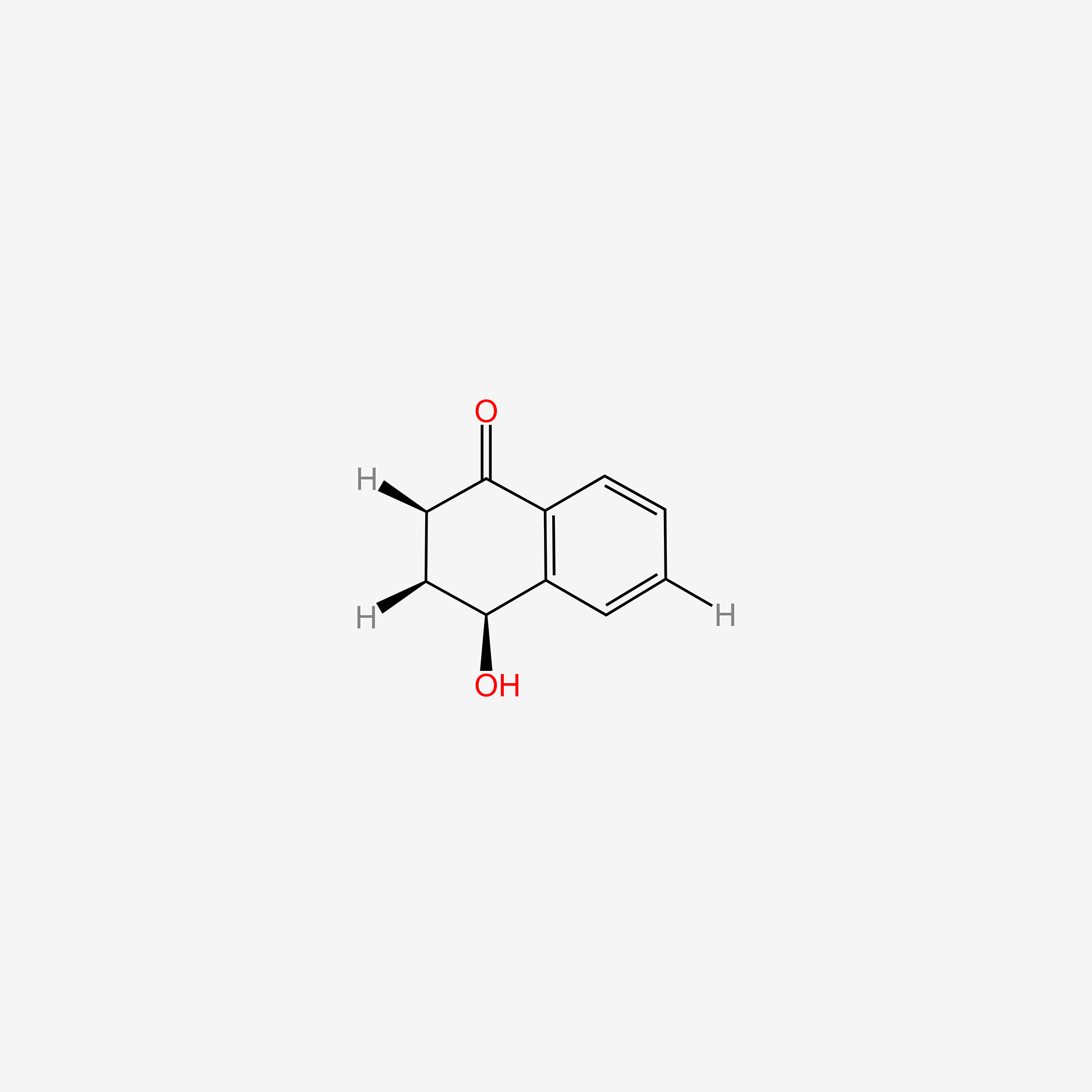 |
0.548 | D03GET |  |
0.367 | ||
| ENC000973 | 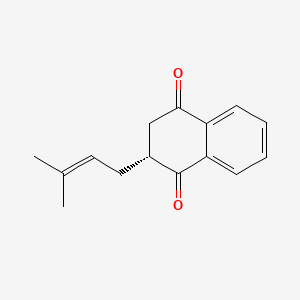 |
0.418 | D06DLI |  |
0.333 | ||
| ENC005244 | 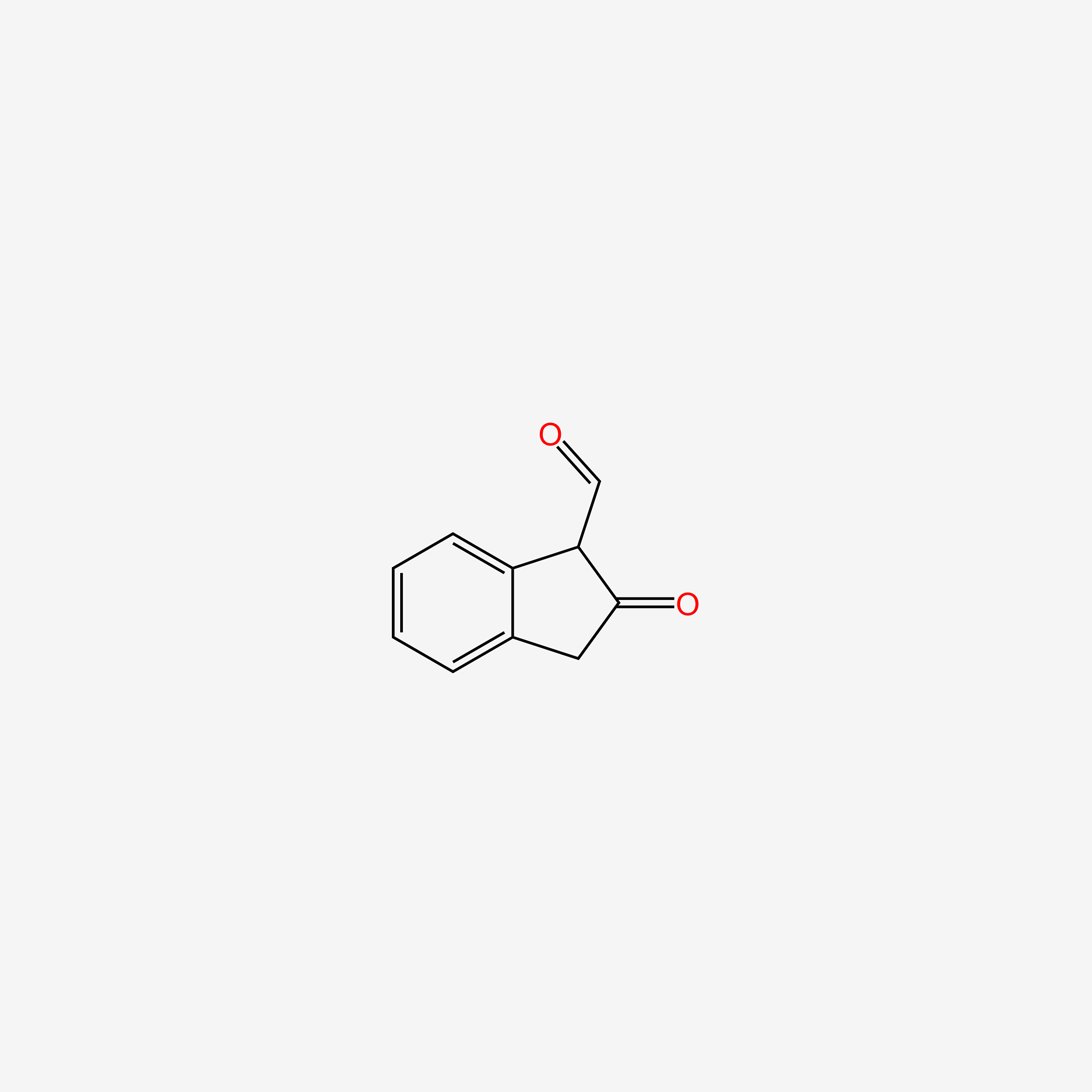 |
0.413 | D0L1WV |  |
0.311 | ||
| ENC000917 |  |
0.405 | D08EOD | 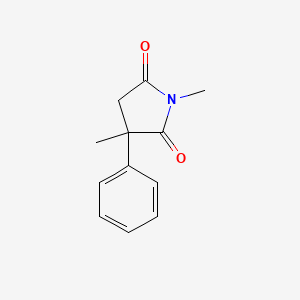 |
0.309 | ||
| ENC000953 | 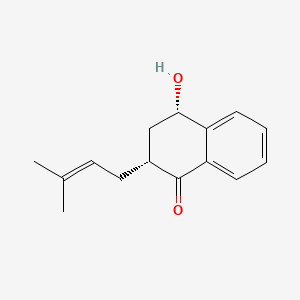 |
0.393 | D0D5GG | 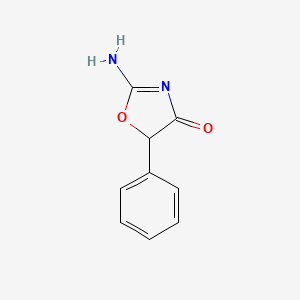 |
0.308 | ||
| ENC004793 | 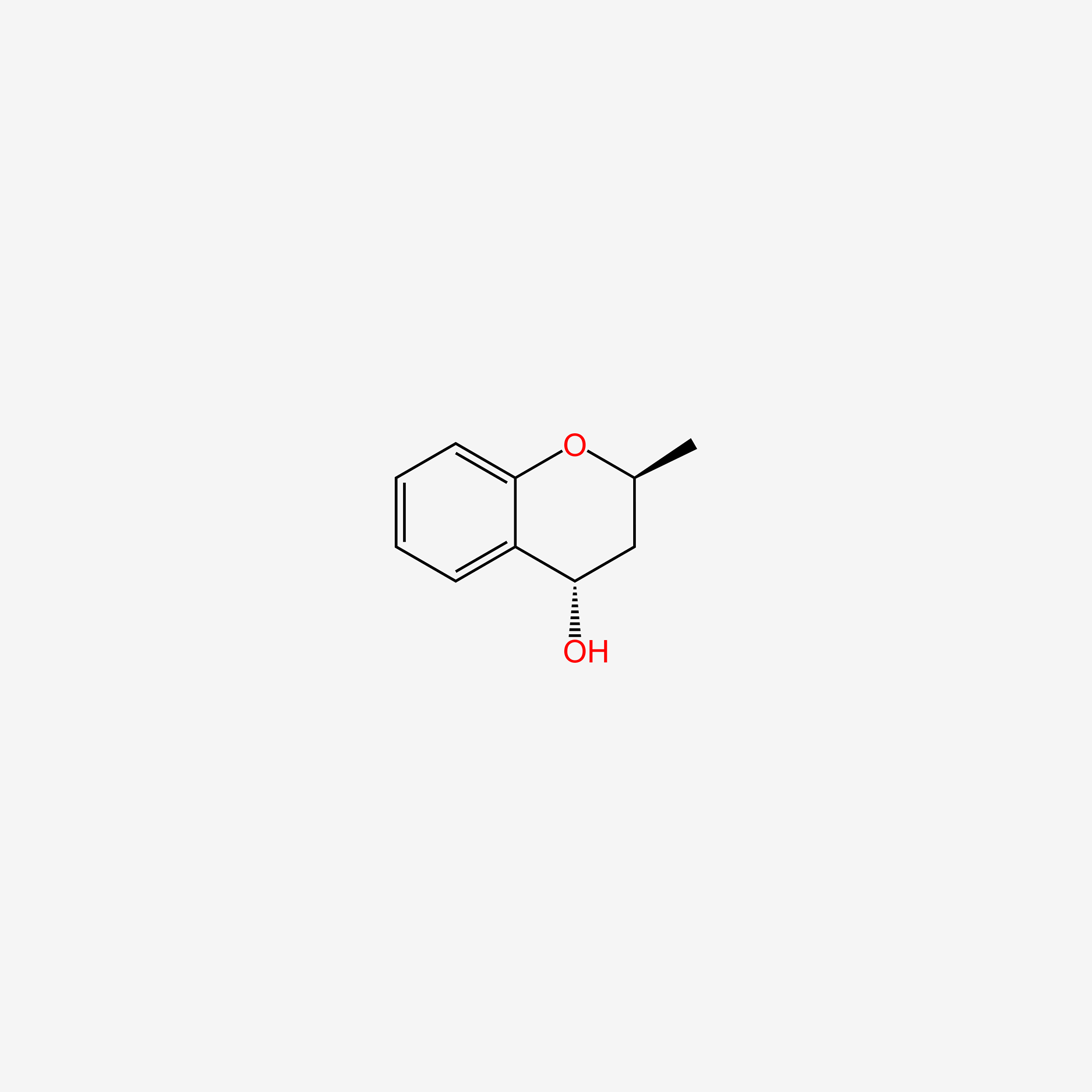 |
0.383 | D05EPM |  |
0.304 | ||
| ENC001319 | 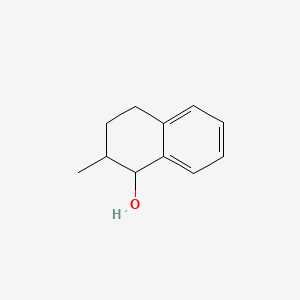 |
0.383 | D06OMW | 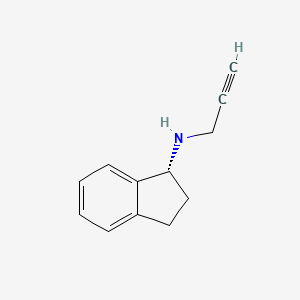 |
0.302 | ||
| ENC005855 | 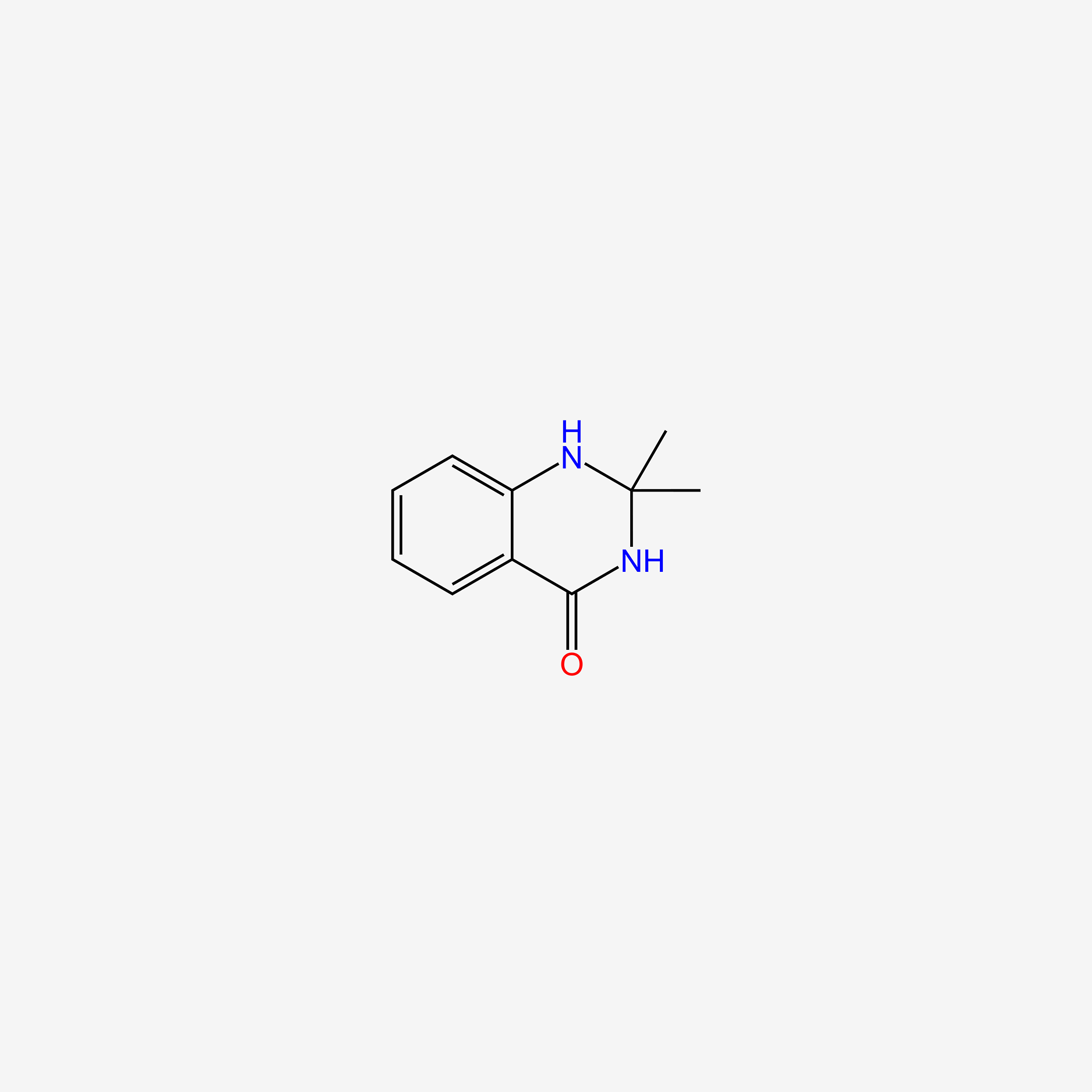 |
0.367 | D0U7GK |  |
0.292 | ||
| ENC000681 | 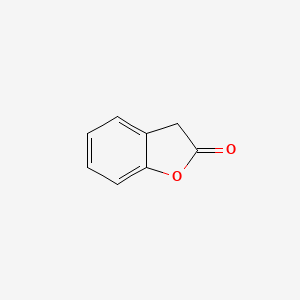 |
0.364 | D02WCI |  |
0.291 | ||