NPs Basic Information
|
Name |
7-Amino-4-methylcoumarin
|
| Molecular Formula | C10H9NO2 | |
| IUPAC Name* |
7-amino-4-methylchromen-2-one
|
|
| SMILES |
CC1=CC(=O)OC2=C1C=CC(=C2)N
|
|
| InChI |
InChI=1S/C10H9NO2/c1-6-4-10(12)13-9-5-7(11)2-3-8(6)9/h2-5H,11H2,1H3
|
|
| InChIKey |
GLNDAGDHSLMOKX-UHFFFAOYSA-N
|
|
| Synonyms |
7-Amino-4-methylcoumarin; 26093-31-2; Coumarin 120; 7-Amino-4-methyl-2H-chromen-2-one; 2H-1-Benzopyran-2-one, 7-amino-4-methyl-; 4-Methyl-7-aminocoumarin; Coumarin, 7-amino-4-methyl-; 7-AMINO-4-METHYL-CHROMEN-2-ONE; 7-amino-4-methylchromen-2-one; AMC; Coumarin 440; 7-amino-4-methyl-2H-1-Benzopyran-2-one; MFCD00006868; OCY3JCT44X; 7-amino-4-methyl coumarin; 7-amino-4-methyl-coumarin; MLS000057660; C10H9NO2; CHEBI:51771; NSC45796; NSC-45796; SMR000067752; (4-Methyl-2-oxo-2H-chromen-7-yl)amine; C 120; CCRIS 4961; EINECS 247-454-8; UNII-OCY3JCT44X; NSC 45796; MCM; NH2Mec; Coumarin 120; AMC; 7-Amino-4-methycoumarin; Maybridge1_002279; 7-amino-4-methylcoumarine; MolMap_000069; 4-methyl-7-amino-coumarin; 7-AMC; Oprea1_663585; SCHEMBL37677; cid_92249; CHEMBL270672; BDBM71742; HMS547P13; ZINC57949; DTXSID40885333; 7-Amino-4-methylcoumarin, 99%; 7-azanyl-4-methyl-chromen-2-one; HMS2343I18; HMS3604G16; HY-D0027; AM9859; HB0694; STK290900; 7-amino-4-methyl-1-benzopyran-2-one; AKOS000308736; CS-W008774; DB08168; PS-6205; SDCCGMLS-0028516.P002; 7-Amino-4-methyl-2H-chromen-2-one #; AC-23087; SY009093; EU-0033321; FT-0621347; FT-0621348; M0760; EN300-05587; A-6591; C01386; 7-Amino-4-methylcoumarin - CAS 26093-31-2; SR-01000597170; J-100007; SR-01000597170-1; 7-Amino-4-methylcoumarin, Chromophore for substrates; Q27097396; Z57024337; 5-{[(4-chlorophenyl)amino]sulfonyl}-3-methyl-N-[3-(trifluoromethyl)benzyl]-1H-pyrazole-4-carboxamide
|
|
| CAS | 26093-31-2 | |
| PubChem CID | 92249 | |
| ChEMBL ID | CHEMBL270672 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 175.18 | ALogp: | 1.6 |
| HBD: | 1 | HBA: | 3 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 52.3 | Aromatic Rings: | 2 |
| Heavy Atoms: | 13 | QED Weighted: | 0.493 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.801 | MDCK Permeability: | 0.00002540 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.999 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.79 |
| 30% Bioavailability (F30%): | 0.997 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.279 | Plasma Protein Binding (PPB): | 77.88% |
| Volume Distribution (VD): | 0.987 | Fu: | 32.31% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.957 | CYP1A2-substrate: | 0.885 |
| CYP2C19-inhibitor: | 0.392 | CYP2C19-substrate: | 0.169 |
| CYP2C9-inhibitor: | 0.167 | CYP2C9-substrate: | 0.4 |
| CYP2D6-inhibitor: | 0.07 | CYP2D6-substrate: | 0.856 |
| CYP3A4-inhibitor: | 0.252 | CYP3A4-substrate: | 0.228 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.103 | Half-life (T1/2): | 0.334 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.07 | Human Hepatotoxicity (H-HT): | 0.172 |
| Drug-inuced Liver Injury (DILI): | 0.728 | AMES Toxicity: | 0.309 |
| Rat Oral Acute Toxicity: | 0.488 | Maximum Recommended Daily Dose: | 0.425 |
| Skin Sensitization: | 0.736 | Carcinogencity: | 0.778 |
| Eye Corrosion: | 0.459 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.639 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001539 |  |
0.714 | D0FA2O |  |
0.361 | ||
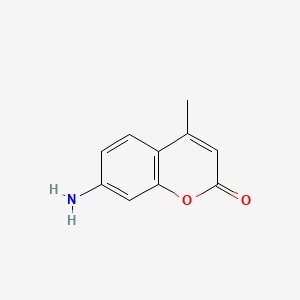
| ENC001393 | 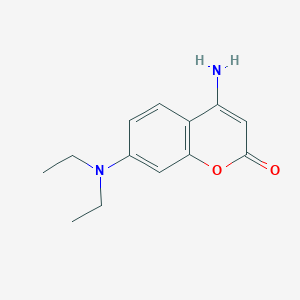 |
0.482 | D0C4YC | 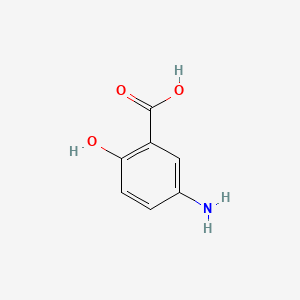 |
0.300 | ||
| ENC002806 | 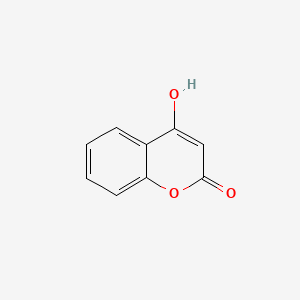 |
0.400 | D01WJL | 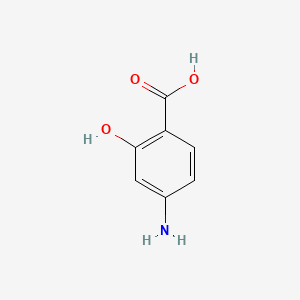 |
0.300 | ||
| ENC000078 | 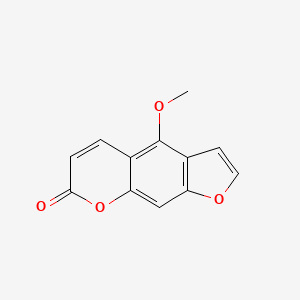 |
0.390 | D0G5UB | 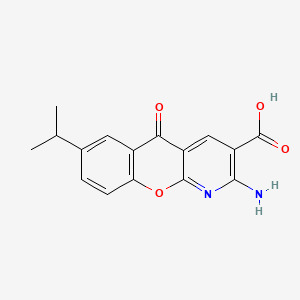 |
0.297 | ||
| ENC001537 |  |
0.389 | D0R9OH |  |
0.297 | ||
| ENC001561 | 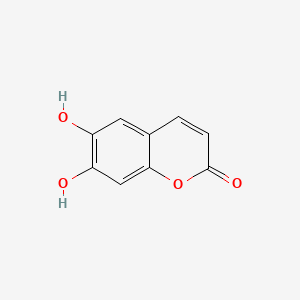 |
0.385 | D0S2BT | 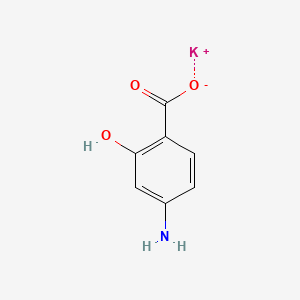 |
0.294 | ||
| ENC001447 |  |
0.358 | D08ZEB |  |
0.288 | ||
| ENC000982 | 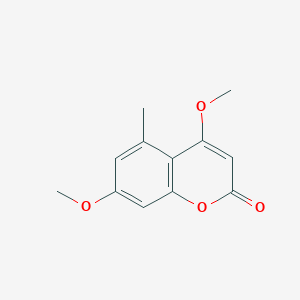 |
0.356 | D09TBD |  |
0.288 | ||
| ENC003861 | 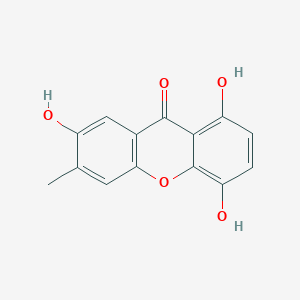 |
0.354 | D08GSF | 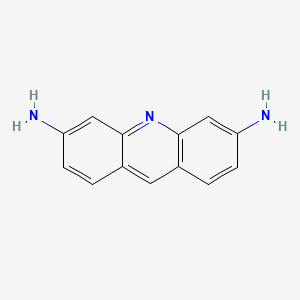 |
0.281 | ||
| ENC001381 | 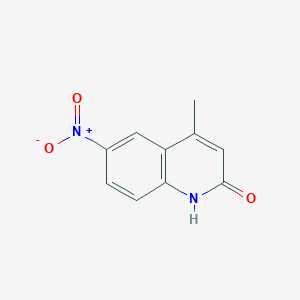 |
0.351 | D08SKH |  |
0.281 | ||