NPs Basic Information
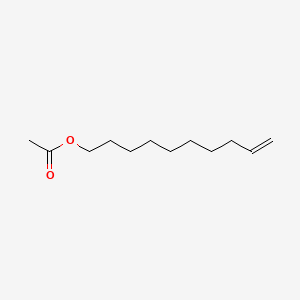
|
Name |
9-Decenyl acetate
|
| Molecular Formula | C12H22O2 | |
| IUPAC Name* |
dec-9-enyl acetate
|
|
| SMILES |
CC(=O)OCCCCCCCCC=C
|
|
| InChI |
InChI=1S/C12H22O2/c1-3-4-5-6-7-8-9-10-11-14-12(2)13/h3H,1,4-11H2,2H3
|
|
| InChIKey |
PIQXMYAEJSMANF-UHFFFAOYSA-N
|
|
| Synonyms |
9-Decenyl acetate; 50816-18-7; 9-Decen-1-ol, acetate; 9-Decen-1-yl acetate; Acetic acid 9-decen-1-yl ester; Decenyl acetate; dec-9-enyl acetate; 9-Decen-1-ol, 1-acetate; 10-Acetoxy-1-decene; dec-9-en-1-yl acetate; Acetic acid 9-decenyl ester; I999R8F793; EINECS 256-784-1; BRN 1765355; AI3-34397; UNII-I999R8F793; ACETICACID9-DECEN-1-YLESTER; ACETIC ACID, 9-DECENYL ESTER; Acetic acid 9-decenyl; DSSTox_CID_27125; DSSTox_RID_82132; 1-ACETOXY-9-DECENE; DSSTox_GSID_47125; 4-02-00-00195 (Beilstein Handbook Reference); SCHEMBL585734; 9-DECEN-1-OL ACETATE; CHEMBL3185404; DTXSID4047125; ZINC2005649; Tox21_302671; MFCD00036512; AKOS024264142; NCGC00256826-01; CAS-50816-18-7; A1093; CS-0453149; FT-0738385; D88376; Q27280605
|
|
| CAS | 50816-18-7 | |
| PubChem CID | 39801 | |
| ChEMBL ID | CHEMBL3185404 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 198.3 | ALogp: | 4.0 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 10 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 14 | QED Weighted: | 0.315 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.599 | MDCK Permeability: | 0.00002880 |
| Pgp-inhibitor: | 0.005 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.735 |
| 30% Bioavailability (F30%): | 0.953 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.998 | Plasma Protein Binding (PPB): | 86.46% |
| Volume Distribution (VD): | 1.201 | Fu: | 27.53% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.966 | CYP1A2-substrate: | 0.276 |
| CYP2C19-inhibitor: | 0.726 | CYP2C19-substrate: | 0.173 |
| CYP2C9-inhibitor: | 0.405 | CYP2C9-substrate: | 0.78 |
| CYP2D6-inhibitor: | 0.106 | CYP2D6-substrate: | 0.237 |
| CYP3A4-inhibitor: | 0.528 | CYP3A4-substrate: | 0.175 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 3.687 | Half-life (T1/2): | 0.504 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.044 | Human Hepatotoxicity (H-HT): | 0.014 |
| Drug-inuced Liver Injury (DILI): | 0.065 | AMES Toxicity: | 0.02 |
| Rat Oral Acute Toxicity: | 0.039 | Maximum Recommended Daily Dose: | 0.015 |
| Skin Sensitization: | 0.955 | Carcinogencity: | 0.36 |
| Eye Corrosion: | 0.981 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.441 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001205 |  |
0.773 | D0Z5BC |  |
0.630 | ||
| ENC001152 |  |
0.765 | D05ATI |  |
0.365 | ||
| ENC000259 |  |
0.660 | D0AY9Q |  |
0.350 | ||
| ENC000455 |  |
0.628 | D0Y8DP |  |
0.333 | ||
| ENC000494 | 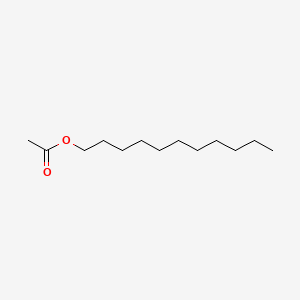 |
0.620 | D0Z5SM | 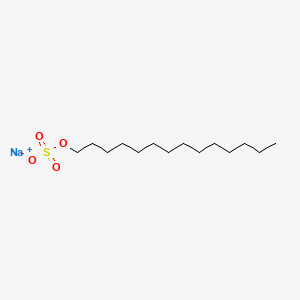 |
0.329 | ||
| ENC000273 |  |
0.587 | D0E4WR |  |
0.321 | ||
| ENC001675 |  |
0.576 | D0O1PH |  |
0.313 | ||
| ENC000647 | 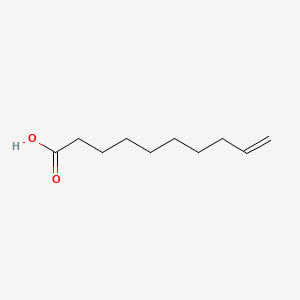 |
0.565 | D0G2KD |  |
0.312 | ||
| ENC000460 | 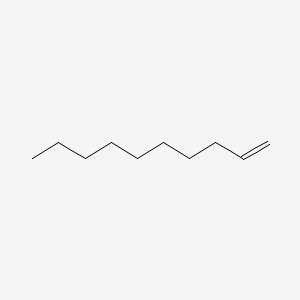 |
0.558 | D0O1TC |  |
0.291 | ||
| ENC000510 |  |
0.551 | D03ZJE |  |
0.289 | ||