NPs Basic Information
|
Name |
1,1,3-Triethoxypropane
|
| Molecular Formula | C9H20O3 | |
| IUPAC Name* |
1,1,3-triethoxypropane
|
|
| SMILES |
CCOCCC(OCC)OCC
|
|
| InChI |
InChI=1S/C9H20O3/c1-4-10-8-7-9(11-5-2)12-6-3/h9H,4-8H2,1-3H3
|
|
| InChIKey |
LGICWIVABSMSDK-UHFFFAOYSA-N
|
|
| Synonyms |
1,1,3-Triethoxypropane; 7789-92-6; 3-Ethoxypropionaldehyde diethyl acetal; Propane, 1,1,3-triethoxy-; 1,3,3-Triethoxypropane; 3-Ethoxypropanal diethyl acetal; 3-Ethoxypropionaldehydediethylacetal; PROPANE, 1,3,3-TRIETHOXY-; Propionaldehyde, 3-ethoxy-, diethyl acetal; .beta.-Ethoxypropionaldehyde diethyl acetal; 1QW8J6VXAM; 1,1,3-triethoxy-propane; NSC-71868; EINECS 232-193-4; UNII-1QW8J6VXAM; NSC 71868; BRN 1098506; beta-Ethoxypropionaldehyde diethyl acetal; AI3-06431; 1,3-Triethoxypropane; Propane,3,3-triethoxy-; SCHEMBL1701153; WLN: 2OYO2&2O2; AMY2529; DTXSID40228509; NSC71868; ZINC1697297; 3-ethoxypropanaldehyde diethyl acetal; MFCD00009241; AKOS009156707; 3-Ethoxypro pionaldehyde diethyl acetal; AS-77291; DB-056268; 3-Ethoxypropionaldehyde diethyl acetal, 95%; FT-0606011; 3-Ethoxypropionaldehyde diethyl acetal, >=95%; D95701; A839282; Q27252769
|
|
| CAS | 7789-92-6 | |
| PubChem CID | 24624 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 176.25 | ALogp: | 1.4 |
| HBD: | 0 | HBA: | 3 |
| Rotatable Bonds: | 8 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 27.7 | Aromatic Rings: | 0 |
| Heavy Atoms: | 12 | QED Weighted: | 0.42 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.065 | MDCK Permeability: | 0.00004570 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.001 | 20% Bioavailability (F20%): | 0.018 |
| 30% Bioavailability (F30%): | 0.007 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.444 | Plasma Protein Binding (PPB): | 22.19% |
| Volume Distribution (VD): | 1.241 | Fu: | 60.05% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.027 | CYP1A2-substrate: | 0.8 |
| CYP2C19-inhibitor: | 0.027 | CYP2C19-substrate: | 0.85 |
| CYP2C9-inhibitor: | 0.008 | CYP2C9-substrate: | 0.071 |
| CYP2D6-inhibitor: | 0.004 | CYP2D6-substrate: | 0.122 |
| CYP3A4-inhibitor: | 0.006 | CYP3A4-substrate: | 0.226 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.189 | Half-life (T1/2): | 0.644 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.101 | Human Hepatotoxicity (H-HT): | 0.027 |
| Drug-inuced Liver Injury (DILI): | 0.022 | AMES Toxicity: | 0.148 |
| Rat Oral Acute Toxicity: | 0.004 | Maximum Recommended Daily Dose: | 0.011 |
| Skin Sensitization: | 0.283 | Carcinogencity: | 0.444 |
| Eye Corrosion: | 0.044 | Eye Irritation: | 0.911 |
| Respiratory Toxicity: | 0.008 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
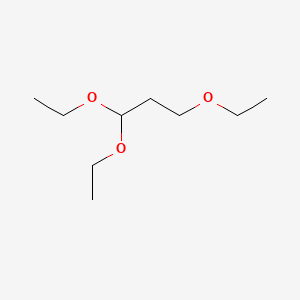
| ENC000547 |  |
0.583 | D0U8AT | 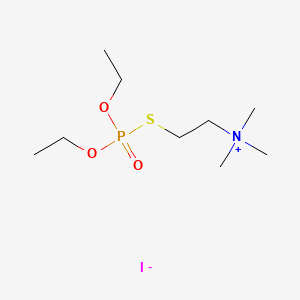 |
0.217 | ||
| ENC000776 | 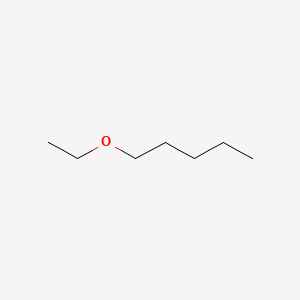 |
0.375 | D0Y3KG |  |
0.204 | ||
| ENC000506 |  |
0.267 | D0K3LW |  |
0.203 | ||
| ENC003057 | 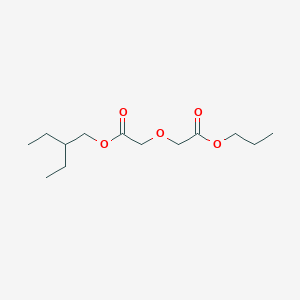 |
0.262 | D0S1ZB |  |
0.203 | ||
| ENC000643 |  |
0.256 | D0K3ZR |  |
0.183 | ||
| ENC000778 | 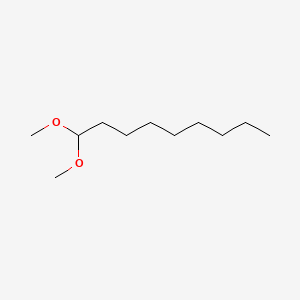 |
0.255 | D0Q7ZG |  |
0.182 | ||
| ENC003080 | 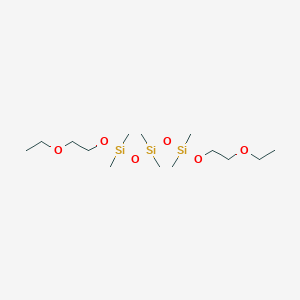 |
0.253 | D0N6CR |  |
0.182 | ||
| ENC000718 |  |
0.250 | D03LGY | 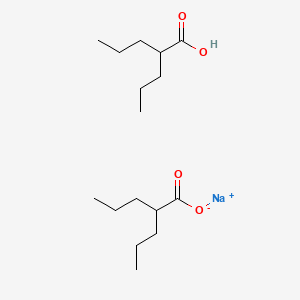 |
0.178 | ||
| ENC000580 | 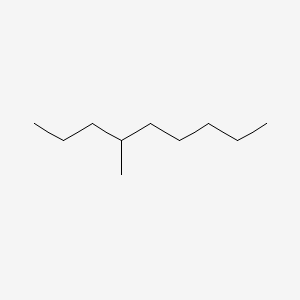 |
0.250 | D0H2SY | 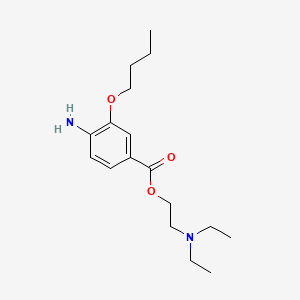 |
0.175 | ||
| ENC000554 |  |
0.250 | D06QDR |  |
0.175 | ||