NPs Basic Information

|
Name |
S-Methyl methanethiosulfonate
|
| Molecular Formula | C2H6O2S2 | |
| IUPAC Name* |
methylsulfonylsulfanylmethane
|
|
| SMILES |
CSS(=O)(=O)C
|
|
| InChI |
InChI=1S/C2H6O2S2/c1-5-6(2,3)4/h1-2H3
|
|
| InChIKey |
XYONNSVDNIRXKZ-UHFFFAOYSA-N
|
|
| Synonyms |
S-Methyl methanethiosulfonate; 2949-92-0; Methyl methanethiolsulfonate; S-Methyl methanesulfonothioate; Methyl methanethiosulfonate; Methanesulfonothioic acid, S-methyl ester; s-methyl methanethiolsulfonate; S-Methyl methanethiosulphonate; Methanethiosulfonic Acid S-Methyl Ester; methylsulfonylsulfanylmethane; Methyl methanesulfonothioate; (METHANESULFONYLSULFANYL)METHANE; s-methyl methylthiosulfonate; dimethyl thiosulfonate; METHANESULFONIC ACID, THIO-, S-METHYL ESTER; methylmethanethiosulfonate; methyl methanethiolsulfate; CHEBI:74357; NSC21545; NSC-21545; Methanethiosulfonic acid, S-methyl ester; 0906Z2356U; S-Methyl thiomethanesulfonate; CCRIS 7217; EINECS 220-970-0; NSC 21545; s-methylmethanethiosulfonate; BRN 1446059; METHYL METHANETHIO-SULFONATE; UNII-0906Z2356U; MeSSO2Me; MFCD00007565; methyl methanethiol sulfonate; methyl methanethiol-sulfonate; methylsulfonylsulfanyl-methane; s-methyl methanethionsulfonate; 4-04-00-00031 (Beilstein Handbook Reference); SCHEMBL136797; CHEMBL1236925; DTXSID8062739; ZINC4268917; S-Methyl methanethiosulfonate, 97%; BBL104000; STL557811; AKOS006221195; CS-W004461; METHYL METHANETHIOSULFONATE, S-; METHYL METHANETHIOSULPHONATE, S-; MS-20744; A5499; AM20080910; FT-0600785; FT-0671958; FT-0671959; M1382; EN300-132006; H10854; S-Methyl methanethiosulfonate, analytical standard; J-017549; Q27144634; S-Methyl methanethiosulfonate, purum, >=98.0% (GC); Methyl methanethiolsulfonate;S-Methyl methanethiolsulfonate
|
|
| CAS | 2949-92-0 | |
| PubChem CID | 18064 | |
| ChEMBL ID | CHEMBL1236925 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 126.2 | ALogp: | 0.0 |
| HBD: | 0 | HBA: | 3 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 67.8 | Aromatic Rings: | 0 |
| Heavy Atoms: | 6 | QED Weighted: | 0.485 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.179 | MDCK Permeability: | 0.00001860 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.008 |
| Human Intestinal Absorption (HIA): | 0.011 | 20% Bioavailability (F20%): | 0.001 |
| 30% Bioavailability (F30%): | 0.009 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.993 | Plasma Protein Binding (PPB): | 24.27% |
| Volume Distribution (VD): | 0.642 | Fu: | 83.80% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.115 | CYP1A2-substrate: | 0.927 |
| CYP2C19-inhibitor: | 0.026 | CYP2C19-substrate: | 0.809 |
| CYP2C9-inhibitor: | 0.014 | CYP2C9-substrate: | 0.19 |
| CYP2D6-inhibitor: | 0.003 | CYP2D6-substrate: | 0.2 |
| CYP3A4-inhibitor: | 0.004 | CYP3A4-substrate: | 0.52 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.469 | Half-life (T1/2): | 0.665 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.015 | Human Hepatotoxicity (H-HT): | 0.359 |
| Drug-inuced Liver Injury (DILI): | 0.537 | AMES Toxicity: | 0.19 |
| Rat Oral Acute Toxicity: | 0.929 | Maximum Recommended Daily Dose: | 0.063 |
| Skin Sensitization: | 0.858 | Carcinogencity: | 0.547 |
| Eye Corrosion: | 0.906 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.88 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000132 |  |
0.533 | D02LDN |  |
0.263 | ||
| ENC000713 | 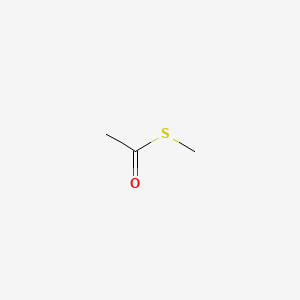 |
0.263 | D07CEI |  |
0.263 | ||
| ENC000417 |  |
0.222 | D08OKJ |  |
0.263 | ||
| ENC000656 | 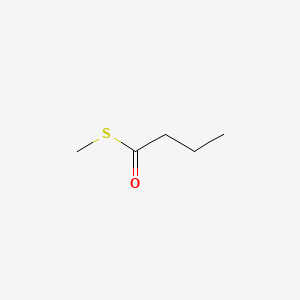 |
0.192 | D0T5DE |  |
0.263 | ||
| ENC000531 |  |
0.182 | D0VB3Y | 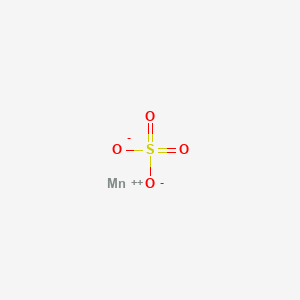 |
0.263 | ||
| ENC000524 |  |
0.160 | D0BG4W | 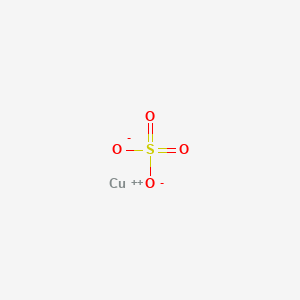 |
0.263 | ||
| ENC000743 | 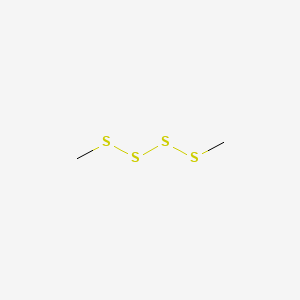 |
0.160 | D07SUG |  |
0.195 | ||
| ENC000418 |  |
0.154 | D0C3YQ |  |
0.192 | ||
| ENC005488 |  |
0.154 | D04YPN | 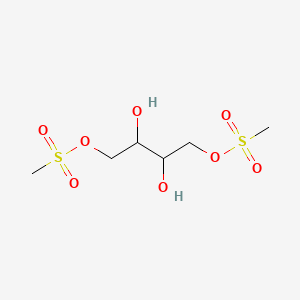 |
0.178 | ||
| ENC000403 | 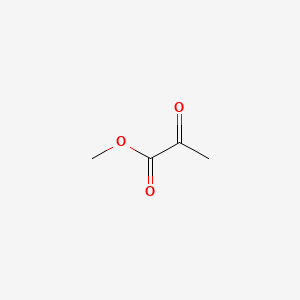 |
0.154 | D0GC2M | 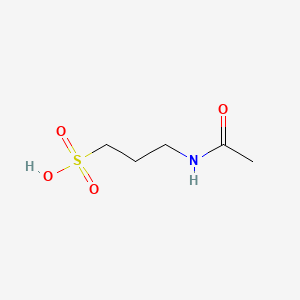 |
0.171 | ||