NPs Basic Information

|
Name |
Dimethyl sulfone
|
| Molecular Formula | C2H6O2S | |
| IUPAC Name* |
methylsulfonylmethane
|
|
| SMILES |
CS(=O)(=O)C
|
|
| InChI |
InChI=1S/C2H6O2S/c1-5(2,3)4/h1-2H3
|
|
| InChIKey |
HHVIBTZHLRERCL-UHFFFAOYSA-N
|
|
| Synonyms |
Dimethyl sulfone; Methyl sulfone; 67-71-0; Methylsulfonylmethane; DIMETHYLSULFONE; Dimethyl sulphone; Sulfonylbismethane; Methane, sulfonylbis-; sulfonyldimethane; Sulphonylbismethane; DMSO2; Methylsulfonyl methane; METHANESULFONYLMETHANE; (methylsulfonyl)methane; METHYL SULFONYL METHANE; NSC 63345; 9H4PO4Z4FT; CHEMBL25028; Methane, 1,1'-sulfonylbis-; CHEBI:9349; NSC-63345; methylsulfone; CCRIS 2938; EINECS 200-665-9; UNII-9H4PO4Z4FT; dimethylsulfon; dimethylsulphone; methy sulfone; methyl sulphone; AI3-25306; Lignisul MSM; Sulfonylbis-methane; Opti MSM; Sulfone, dimethyl-; MFCD00007566; (methylsulphonyl)methane; Dimethyl sulfone, 98%; METHOSULFONYLMETHANE; 2-Thiapropane2,2-dioxide; MolMap_000019; DSSTox_CID_23937; DSSTox_GSID_43937; DIMETHYL SULFONE [MI]; SPECTRUM1505358; DIMETHYL SULFONE [INCI]; DTXSID4043937; DIMETHYL SULFONE [MART.]; AMY25756; HY-Y1314; NSC63345; ZINC4658606; METHYLSULFONYLMETHANE [VANDF]; Tox21_303712; BDBM50026473; METHYLSULFONYLMETHANE [USP-RS]; METHYLSULFONYLMETHANE [WHO-DD]; AKOS015897615; CCG-214558; DB14090; CAS-67-71-0; NCGC00095990-01; NCGC00357027-01; DB-050533; MYTHYLSULFONYLMETHANE (MSM) [VANDF]; CS-0017786; FT-0625160; M0509; M1239; EN300-79559; D70240; A835859; Q423842; DIMETHYL SULFOXIDE IMPURITY A [EP IMPURITY]; F0001-1776; Z417007936; Dimethyl sulfone, Standard for quantitative NMR, TraceCERT(R); Methylsulfonylmethane, United States Pharmacopeia (USP) Reference Standard; Methylsulfonylmethane, Pharmaceutical Secondary Standard; Certified Reference Material
|
|
| CAS | 67-71-0 | |
| PubChem CID | 6213 | |
| ChEMBL ID | CHEMBL25028 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 94.14 | ALogp: | -0.4 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 42.5 | Aromatic Rings: | 0 |
| Heavy Atoms: | 5 | QED Weighted: | 0.426 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.341 | MDCK Permeability: | 0.00001770 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.006 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.001 |
| 30% Bioavailability (F30%): | 0.002 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.966 | Plasma Protein Binding (PPB): | 9.92% |
| Volume Distribution (VD): | 0.581 | Fu: | 88.97% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.281 | CYP1A2-substrate: | 0.969 |
| CYP2C19-inhibitor: | 0.014 | CYP2C19-substrate: | 0.738 |
| CYP2C9-inhibitor: | 0.002 | CYP2C9-substrate: | 0.067 |
| CYP2D6-inhibitor: | 0.002 | CYP2D6-substrate: | 0.128 |
| CYP3A4-inhibitor: | 0.004 | CYP3A4-substrate: | 0.68 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 1.208 | Half-life (T1/2): | 0.611 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.007 | Human Hepatotoxicity (H-HT): | 0.228 |
| Drug-inuced Liver Injury (DILI): | 0.039 | AMES Toxicity: | 0.003 |
| Rat Oral Acute Toxicity: | 0.009 | Maximum Recommended Daily Dose: | 0.038 |
| Skin Sensitization: | 0.567 | Carcinogencity: | 0.334 |
| Eye Corrosion: | 0.012 | Eye Irritation: | 0.942 |
| Respiratory Toxicity: | 0.056 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000522 |  |
0.533 | D0T5DE |  |
0.313 | ||
| ENC000568 |  |
0.176 | D08OKJ |  |
0.313 | ||
| ENC000403 | 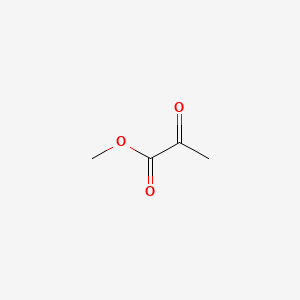 |
0.174 | D0VB3Y | 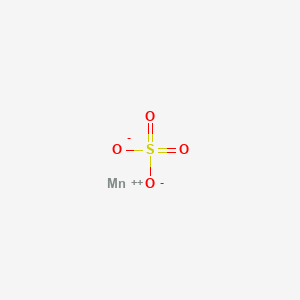 |
0.313 | ||
| ENC005488 |  |
0.174 | D02LDN |  |
0.313 | ||
| ENC000418 |  |
0.174 | D0BG4W | 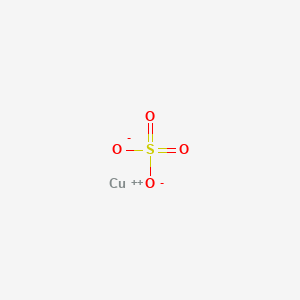 |
0.313 | ||
| ENC000135 | 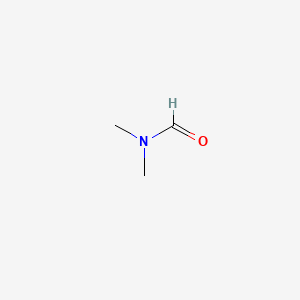 |
0.167 | D07CEI |  |
0.313 | ||
| ENC000713 | 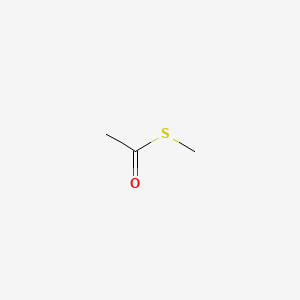 |
0.167 | D07SUG |  |
0.243 | ||
| ENC000410 | 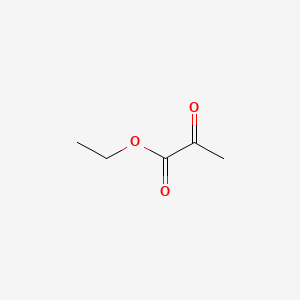 |
0.154 | D04YPN | 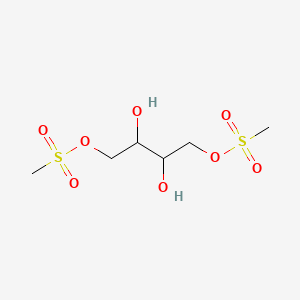 |
0.220 | ||
| ENC000682 | 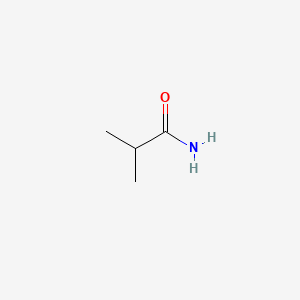 |
0.150 | D0C3YQ |  |
0.217 | ||
| ENC000010 |  |
0.150 | D08HVE |  |
0.200 | ||