NPs Basic Information

|
Name |
alpha-Thujene
|
| Molecular Formula | C10H16 | |
| IUPAC Name* |
2-methyl-5-propan-2-ylbicyclo[3.1.0]hex-2-ene
|
|
| SMILES |
CC1=CCC2(C1C2)C(C)C
|
|
| InChI |
InChI=1S/C10H16/c1-7(2)10-5-4-8(3)9(10)6-10/h4,7,9H,5-6H2,1-3H3
|
|
| InChIKey |
KQAZVFVOEIRWHN-UHFFFAOYSA-N
|
|
| Synonyms |
ALPHA-THUJENE; 3-Thujene; 2867-05-2; Origanene; Bicyclo[3.1.0]hex-2-ene, 2-methyl-5-(1-methylethyl)-; 2-methyl-5-propan-2-ylbicyclo[3.1.0]hex-2-ene; thujiene; 2-methyl-5-(propan-2-yl)bicyclo[3.1.0]hex-2-ene; .alpha.-Thujene; 5-Isopropyl-2-methylbicyclo[3.1.0]hex-2-ene; 2-Methyl-5-(1-methylethyl)-bicyclo(3.1.0)hex-2-ene; 2-methyl-5-(1-methylethyl)-bicyclo[3.1.0]hex-2-ene; alpha-Thuiene; alpha-Thujen; thujene (alpha-); 4-methyl-1-propan-2-ylbicyclo[3.1.0]hex-3-ene; Thujene, .alpha.-; (-)-3-Thujene; Thujone,(a + b)(sg); (1R,5S)-thuj-2-ene; (1R,5S)-2-methyl-5-(propan-2-yl)bicyclo[3.1.0]hex-2-ene; CHEBI:50031; DTXSID20863038; FT-0622205; EN300-7441739; 5-Isopropyl-2-methylbicyclo[3.1.0]hex-2-ene #; Q27121796
|
|
| CAS | 2867-05-2 | |
| PubChem CID | 17868 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 136.23 | ALogp: | 2.8 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 10 | QED Weighted: | 0.481 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.28 | MDCK Permeability: | 0.00002360 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.79 |
| 30% Bioavailability (F30%): | 0.815 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.696 | Plasma Protein Binding (PPB): | 91.48% |
| Volume Distribution (VD): | 1.979 | Fu: | 9.76% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.701 | CYP1A2-substrate: | 0.453 |
| CYP2C19-inhibitor: | 0.267 | CYP2C19-substrate: | 0.914 |
| CYP2C9-inhibitor: | 0.165 | CYP2C9-substrate: | 0.414 |
| CYP2D6-inhibitor: | 0.081 | CYP2D6-substrate: | 0.583 |
| CYP3A4-inhibitor: | 0.234 | CYP3A4-substrate: | 0.289 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 16.534 | Half-life (T1/2): | 0.19 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.014 | Human Hepatotoxicity (H-HT): | 0.24 |
| Drug-inuced Liver Injury (DILI): | 0.124 | AMES Toxicity: | 0.013 |
| Rat Oral Acute Toxicity: | 0.072 | Maximum Recommended Daily Dose: | 0.065 |
| Skin Sensitization: | 0.103 | Carcinogencity: | 0.463 |
| Eye Corrosion: | 0.609 | Eye Irritation: | 0.983 |
| Respiratory Toxicity: | 0.113 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC002844 |  |
0.511 | D0A2AJ |  |
0.203 | ||
| ENC000528 |  |
0.421 | D04CSZ |  |
0.191 | ||
| ENC001637 |  |
0.366 | D06GIP |  |
0.191 | ||
| ENC000388 |  |
0.366 | D0H1QY | 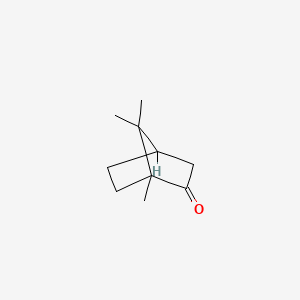 |
0.167 | ||
| ENC000153 |  |
0.350 | D08KVZ |  |
0.164 | ||
| ENC004826 |  |
0.333 | D05VQI | 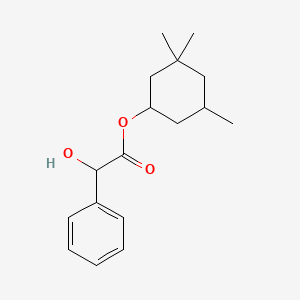 |
0.155 | ||
| ENC002232 |  |
0.333 | D0K7LU | 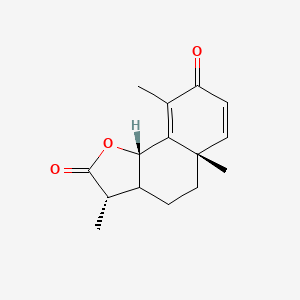 |
0.152 | ||
| ENC002220 |  |
0.333 | D0A3HB |  |
0.151 | ||
| ENC002143 |  |
0.333 | D0X0RI |  |
0.148 | ||
| ENC004827 |  |
0.333 | D0TQ1G |  |
0.143 | ||