NPs Basic Information
|
Name |
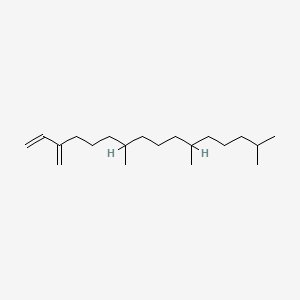
Neophytadiene
|
| Molecular Formula | C20H38 | |
| IUPAC Name* |
7,11,15-trimethyl-3-methylidenehexadec-1-ene
|
|
| SMILES |
CC(C)CCCC(C)CCCC(C)CCCC(=C)C=C
|
|
| InChI |
InChI=1S/C20H38/c1-7-18(4)12-9-14-20(6)16-10-15-19(5)13-8-11-17(2)3/h7,17,19-20H,1,4,8-16H2,2-3,5-6H3
|
|
| InChIKey |
NIDGCIPAMWNKOA-UHFFFAOYSA-N
|
|
| Synonyms |
NEOPHYTADIENE; 504-96-1; 7,11,15-trimethyl-3-methylidenehexadec-1-ene; 2-(4,8,12-Trimethyltridecyl)buta-1,3-diene; 1-Hexadecene, 7,11,15-trimethyl-3-methylene-; 3-Methylene-7,11,15-trimethylhexadec-1-ene; HL3QFB56FB; 1,3-Butadiene, 2-(4,8,12-trimethyltridecyl)-; 3-METHYLENE-7,11,15-TRIMETHYL-1-HEXADECENE; HSDB 4321; Neophytadiene (80%); UNII-HL3QFB56FB; 7,11,15-Trimethyl-3-methylene-1-hexadecene; 2-(4,8,12-Trimethyltridecyl)-1,3-butadiene; Neophytadiene (80per cent); 7,11,15-trimethyl-3-methylidene-hexadec-1-ene; DTXSID40964657; CHEBI:145817; EX-A2765; HY-N8534; AKOS030254489; CS-0145915; FT-0672681; 7,11,15-Trimethyl-3-methylenehexadec-1-ene; Q67880018; 2-(4,8,12-TRIMETHYLTRIDECYL)BUTA-1,3-DIENE [HSDB]
|
|
| CAS | 504-96-1 | |
| PubChem CID | 10446 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 278.5 | ALogp: | 9.6 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 13 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 20 | QED Weighted: | 0.314 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.628 | MDCK Permeability: | 0.00000544 |
| Pgp-inhibitor: | 0.154 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.002 | 20% Bioavailability (F20%): | 0.004 |
| 30% Bioavailability (F30%): | 0.359 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.513 | Plasma Protein Binding (PPB): | 98.55% |
| Volume Distribution (VD): | 3.743 | Fu: | 1.67% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.442 | CYP1A2-substrate: | 0.2 |
| CYP2C19-inhibitor: | 0.352 | CYP2C19-substrate: | 0.557 |
| CYP2C9-inhibitor: | 0.498 | CYP2C9-substrate: | 0.934 |
| CYP2D6-inhibitor: | 0.058 | CYP2D6-substrate: | 0.072 |
| CYP3A4-inhibitor: | 0.349 | CYP3A4-substrate: | 0.186 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.58 | Half-life (T1/2): | 0.043 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.055 |
| Drug-inuced Liver Injury (DILI): | 0.856 | AMES Toxicity: | 0.013 |
| Rat Oral Acute Toxicity: | 0.01 | Maximum Recommended Daily Dose: | 0.08 |
| Skin Sensitization: | 0.965 | Carcinogencity: | 0.082 |
| Eye Corrosion: | 0.977 | Eye Irritation: | 0.938 |
| Respiratory Toxicity: | 0.904 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000766 |  |
0.695 | D00FSV |  |
0.450 | ||
| ENC001722 |  |
0.682 | D0ZI4H | 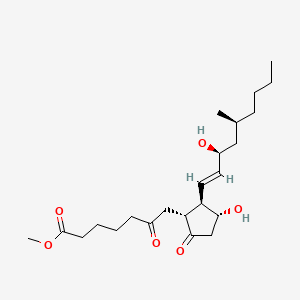 |
0.220 | ||
| ENC000538 | 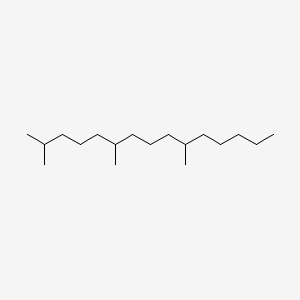 |
0.661 | D0Z5BC |  |
0.216 | ||
| ENC000902 |  |
0.644 | D03LGY | 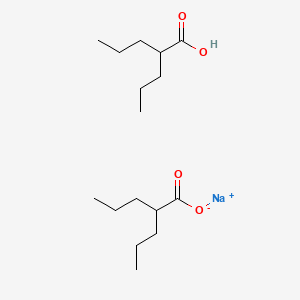 |
0.216 | ||
| ENC000537 |  |
0.644 | D0D9NY |  |
0.206 | ||
| ENC001286 | 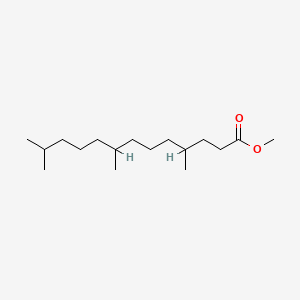 |
0.615 | D0T9TJ |  |
0.197 | ||
| ENC000441 | 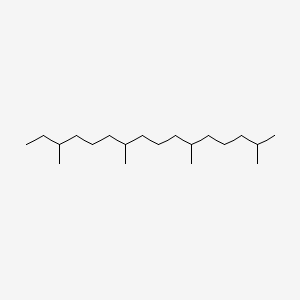 |
0.612 | D0N3NO | 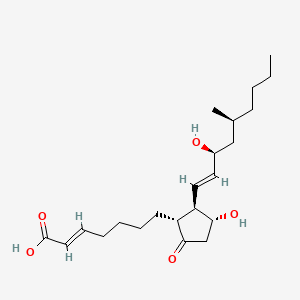 |
0.196 | ||
| ENC001818 | 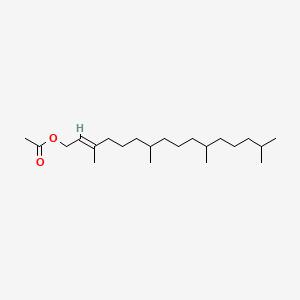 |
0.608 | D0G2KD |  |
0.196 | ||
| ENC001412 | 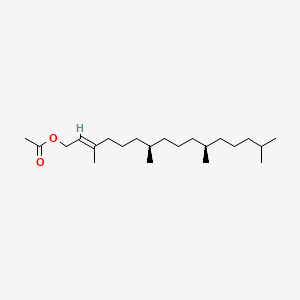 |
0.608 | D0X4FM | 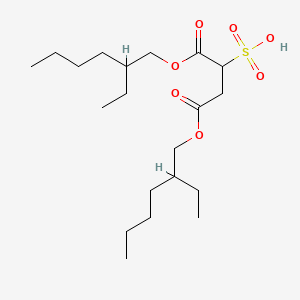 |
0.194 | ||
| ENC000536 | 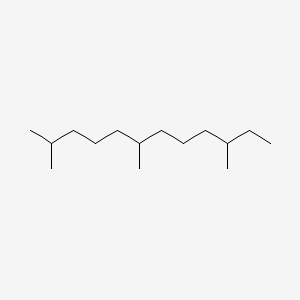 |
0.593 | D0K5WS |  |
0.189 | ||