NPs Basic Information
|
Name |
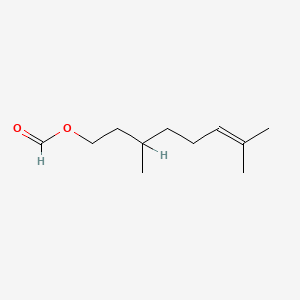
Citronellyl formate
|
| Molecular Formula | C11H20O2 | |
| IUPAC Name* |
3,7-dimethyloct-6-enyl formate
|
|
| SMILES |
CC(CCC=C(C)C)CCOC=O
|
|
| InChI |
InChI=1S/C11H20O2/c1-10(2)5-4-6-11(3)7-8-13-9-12/h5,9,11H,4,6-8H2,1-3H3
|
|
| InChIKey |
DZNVIZQPWLDQHI-UHFFFAOYSA-N
|
|
| Synonyms |
Citronellyl formate; 105-85-1; 3,7-dimethyloct-6-en-1-yl formate; 6-OCTEN-1-OL, 3,7-DIMETHYL-, FORMATE; Citronellol formate; Formic acid, citronellyl ester; Citronellyl methanoate; 2,6-Dimethyl-2-octen-8-yl formate; 3,7-Dimethyl-6-octen-1-yl formate; 3,7-dimethyloct-6-enyl formate; FEMA No. 2314; Formic acid, 3,7-dimethyl-6-octen-1-yl ester; (+)-3,7-Dimethyloct-6-enyl formate; (-)-3,7-Dimethyloct-6-enyl formate; (1)-3,7-Dimethyloct-6-enyl formate; 3,7-Dimethyl-6-octen-1-yl methanoate; 93919-91-6; 7B1MY2BRDK; 6-Octen-1-ol, 3,7-dimethyl-, 1-formate; CHEBI:31406; NSC-46117; 93919-93-8; EINECS 203-338-9; EINECS 300-075-2; UNII-7B1MY2BRDK; NSC 46117; Citronellol formate (6CI); BRN 1723215; AI3-24239; Citronelyl formate; EINECS 300-076-8; EINECS 300-078-9; 6-Octen-1-ol,3,7-dimethyl-, 1-formate; Citronellyl formate, FCC; DSSTox_CID_24772; DSSTox_RID_80463; DSSTox_GSID_44772; Formic acid citronellyl ester; 2-02-00-00032 (Beilstein Handbook Reference); 3,7-Dimethyl-6-octen--yl; SCHEMBL416782; CHEMBL3184928; DTXSID1044772; FEMA 2314; CITRONELLYL FORMATE [FCC]; WLN: VHO2Y1&3UY1&1; CITRONELLYL FORMATE [FHFI]; (+/-)-CITRONELLYL FORMATE; NSC46117; 3,7-Dimethyl-6-octenyl formate #; Tox21_301031; 6-Octen-1-ol,7-dimethyl-, formate; AKOS016009786; 2, 6-Dimethyl-2-octen-8-yl formate; CITRONELLYL FORMATE, (+/-)-; LMPR0102010014; NCGC00248262-01; NCGC00254933-01; 6-Octen-1-ol,3,7-dimethyl-,1-formate; AS-62928; CAS-105-85-1; Formic acid,7-dimethyl-6-octen-1-yl ester; CS-0186751; FT-0623969; (+)-Formic acid 3,7-dimethyl-6-octenyl ester; (?)-Formic acid 3,7-dimethyl-6-octenyl ester; W-109390; Q27114304
|
|
| CAS | 93919-91-6 | |
| PubChem CID | 7778 | |
| ChEMBL ID | CHEMBL3184928 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 184.27 | ALogp: | 3.8 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 7 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 13 | QED Weighted: | 0.342 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.408 | MDCK Permeability: | 0.00002610 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.005 | 20% Bioavailability (F20%): | 0.016 |
| 30% Bioavailability (F30%): | 0.019 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.995 | Plasma Protein Binding (PPB): | 85.18% |
| Volume Distribution (VD): | 2.389 | Fu: | 8.73% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.961 | CYP1A2-substrate: | 0.214 |
| CYP2C19-inhibitor: | 0.525 | CYP2C19-substrate: | 0.569 |
| CYP2C9-inhibitor: | 0.283 | CYP2C9-substrate: | 0.63 |
| CYP2D6-inhibitor: | 0.055 | CYP2D6-substrate: | 0.155 |
| CYP3A4-inhibitor: | 0.078 | CYP3A4-substrate: | 0.186 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.43 | Half-life (T1/2): | 0.357 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.009 | Human Hepatotoxicity (H-HT): | 0.697 |
| Drug-inuced Liver Injury (DILI): | 0.093 | AMES Toxicity: | 0.006 |
| Rat Oral Acute Toxicity: | 0.009 | Maximum Recommended Daily Dose: | 0.03 |
| Skin Sensitization: | 0.935 | Carcinogencity: | 0.441 |
| Eye Corrosion: | 0.664 | Eye Irritation: | 0.93 |
| Respiratory Toxicity: | 0.176 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000319 | 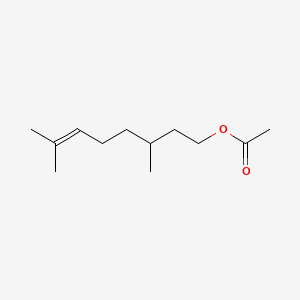 |
0.636 | D0M1PQ |  |
0.419 | ||
| ENC000311 | 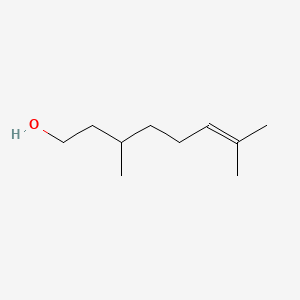 |
0.600 | D0ZK8H |  |
0.222 | ||
| ENC000230 | 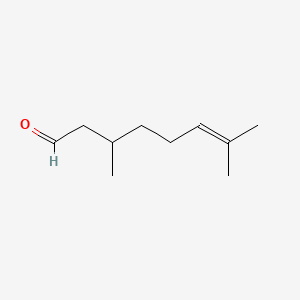 |
0.600 | D0Y3KG |  |
0.220 | ||
| ENC001649 |  |
0.458 | D09XWD |  |
0.220 | ||
| ENC000846 |  |
0.452 | D05XQE |  |
0.218 | ||
| ENC003366 |  |
0.451 | D0Q9HF |  |
0.200 | ||
| ENC001434 |  |
0.362 | D04MWJ |  |
0.192 | ||
| ENC001424 |  |
0.362 | D0U5CE | 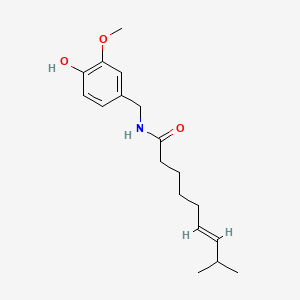 |
0.185 | ||
| ENC001150 | 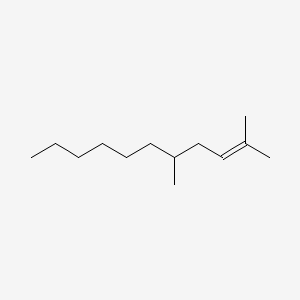 |
0.346 | D03LGG | 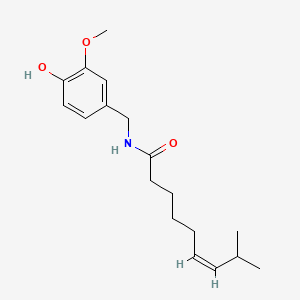 |
0.185 | ||
| ENC000145 |  |
0.333 | D03VFL | 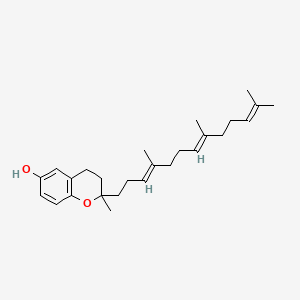 |
0.177 | ||