NPs Basic Information

|
Name |
(S)-3-Benzylpiperazine-2,5-dione
|
| Molecular Formula | C11H12N2O2 | |
| IUPAC Name* |
(3S)-3-benzylpiperazine-2,5-dione
|
|
| SMILES |
C1C(=O)N[C@H](C(=O)N1)CC2=CC=CC=C2
|
|
| InChI |
InChI=1S/C11H12N2O2/c14-10-7-12-11(15)9(13-10)6-8-4-2-1-3-5-8/h1-5,9H,6-7H2,(H,12,15)(H,13,14)/t9-/m0/s1
|
|
| InChIKey |
UZOJHXFWJFSFAI-VIFPVBQESA-N
|
|
| Synonyms |
10125-07-2; CYCLO(-GLY-PHE); (S)-3-BENZYLPIPERAZINE-2,5-DIONE; (3S)-3-benzylpiperazine-2,5-dione; CYCLO(GLY-L-PHE); (S)-3-BENZYL-2,5-DIOXOPIPERAZINE; cyclo(Gly-Phe); 2,5-Piperazinedione, 3-(phenylmethyl)-, (3S)-; (S)-3-Benzyl-piperazine-2,5-dione; Cyclo(-Gly-L-Phe); Cyclo(L-Phe-Gly-); cyclo(glycyl-L-phenylalanyl); cyclo(L-phenylalanyl-glycyl); Cyclo(-Gly-Phe), AldrichCPR; SCHEMBL1673516; CHEMBL2074841; ZINC4026502; (S)-2-benzylpiperazine-3,6-dione; MFCD00190926; (S)-2-benzyl piperazine-3,6-dione; 3-(S)-benzyl-piperazine-2,5-dione; 6-(S)-Benzyl-piperazine-2,5-dione; AKOS024462543; BS-19083; A897211
|
|
| CAS | NA | |
| PubChem CID | 7076549 | |
| ChEMBL ID | CHEMBL2074841 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 204.22 | ALogp: | 0.3 |
| HBD: | 2 | HBA: | 2 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 58.2 | Aromatic Rings: | 2 |
| Heavy Atoms: | 15 | QED Weighted: | 0.726 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.945 | MDCK Permeability: | 0.00005510 |
| Pgp-inhibitor: | 0.001 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.233 | 20% Bioavailability (F20%): | 0.596 |
| 30% Bioavailability (F30%): | 0.027 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.975 | Plasma Protein Binding (PPB): | 32.24% |
| Volume Distribution (VD): | 0.484 | Fu: | 58.86% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.046 | CYP1A2-substrate: | 0.087 |
| CYP2C19-inhibitor: | 0.117 | CYP2C19-substrate: | 0.119 |
| CYP2C9-inhibitor: | 0.053 | CYP2C9-substrate: | 0.138 |
| CYP2D6-inhibitor: | 0.007 | CYP2D6-substrate: | 0.174 |
| CYP3A4-inhibitor: | 0.066 | CYP3A4-substrate: | 0.143 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 5.794 | Half-life (T1/2): | 0.739 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.035 | Human Hepatotoxicity (H-HT): | 0.092 |
| Drug-inuced Liver Injury (DILI): | 0.041 | AMES Toxicity: | 0.071 |
| Rat Oral Acute Toxicity: | 0.151 | Maximum Recommended Daily Dose: | 0.052 |
| Skin Sensitization: | 0.304 | Carcinogencity: | 0.046 |
| Eye Corrosion: | 0.003 | Eye Irritation: | 0.018 |
| Respiratory Toxicity: | 0.049 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001905 | 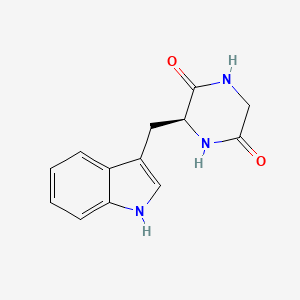 |
0.610 | D0U5RT |  |
0.397 | ||
| ENC002255 |  |
0.576 | D05EPM |  |
0.393 | ||
| ENC001909 | 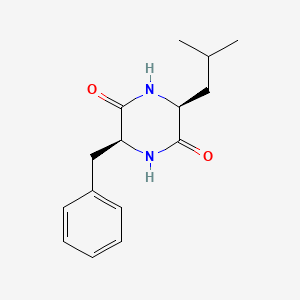 |
0.548 | D03RZV | 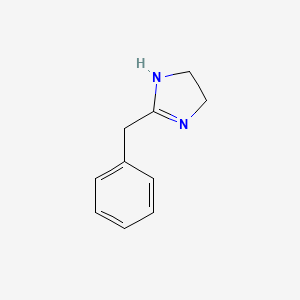 |
0.386 | ||
| ENC001087 | 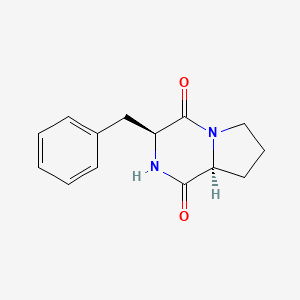 |
0.532 | D0Z9NZ | 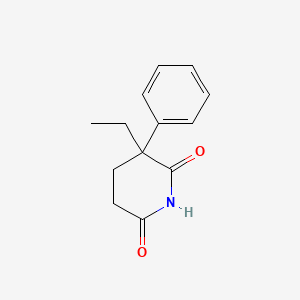 |
0.375 | ||
| ENC005971 |  |
0.532 | D05OIS |  |
0.375 | ||
| ENC005484 | 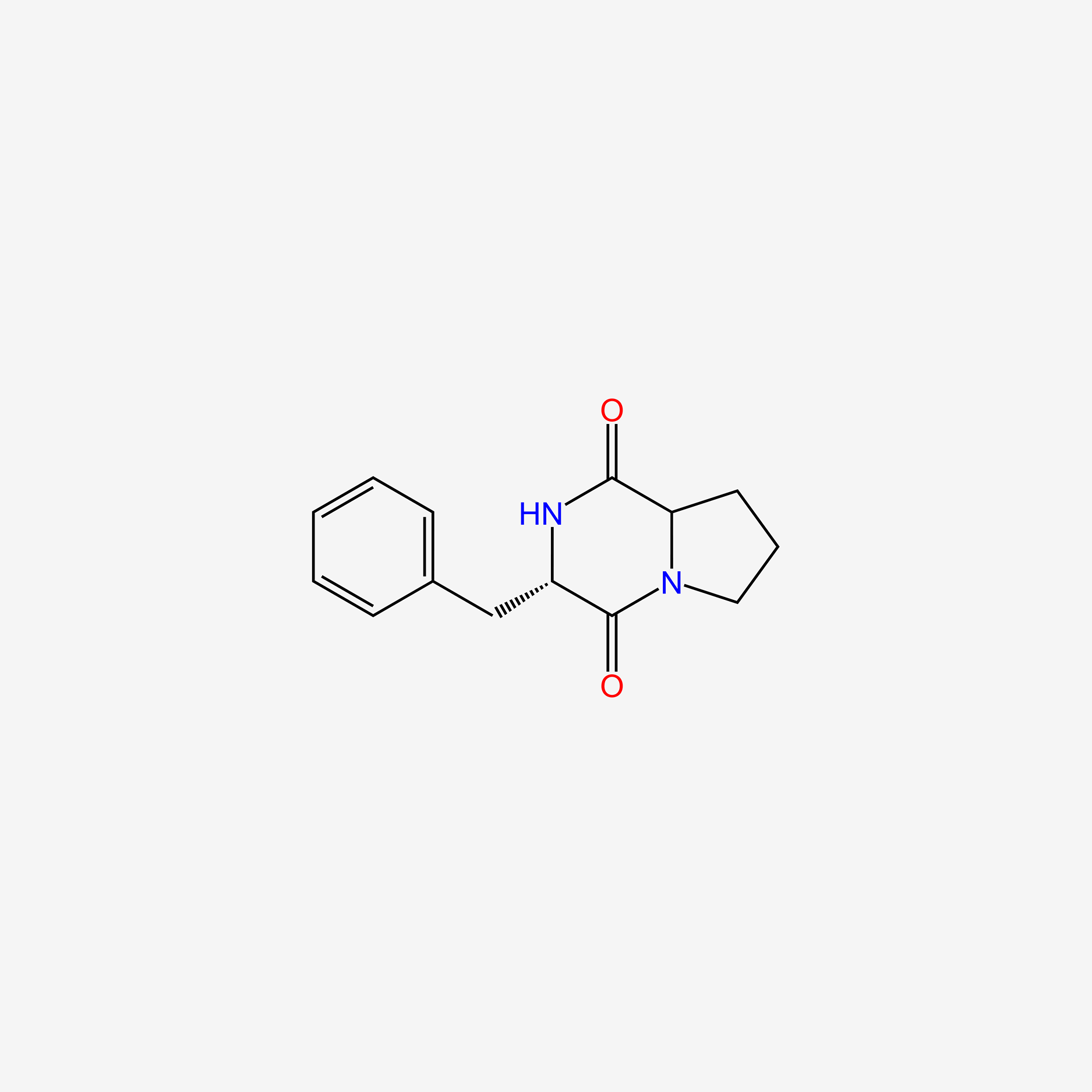 |
0.532 | D06BYV |  |
0.367 | ||
| ENC000825 | 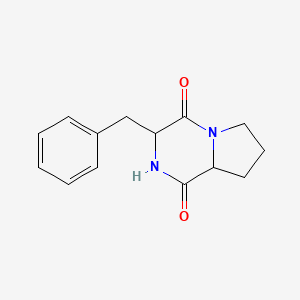 |
0.532 | D0Y7RW |  |
0.343 | ||
| ENC005847 |  |
0.516 | D0T3LF |  |
0.340 | ||
| ENC002030 |  |
0.516 | D05BMG |  |
0.340 | ||
| ENC004648 |  |
0.507 | D07ONP |  |
0.339 | ||