NPs Basic Information
|
Name |
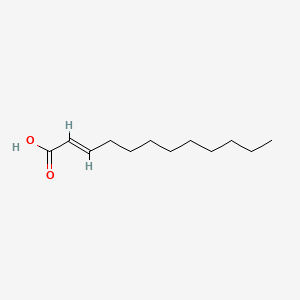
2-Dodecenoic acid
|
| Molecular Formula | C12H22O2 | |
| IUPAC Name* |
(E)-dodec-2-enoic acid
|
|
| SMILES |
CCCCCCCCC/C=C/C(=O)O
|
|
| InChI |
InChI=1S/C12H22O2/c1-2-3-4-5-6-7-8-9-10-11-12(13)14/h10-11H,2-9H2,1H3,(H,13,14)/b11-10+
|
|
| InChIKey |
PAWGRNGPMLVJQH-ZHACJKMWSA-N
|
|
| Synonyms |
2-Dodecenoic acid; Dodec-2-enoic acid; (E)-dodec-2-enoic acid; trans-2-Dodecenoic acid; 32466-54-9; 4412-16-2; (2E)-dodec-2-enoic acid; (2E)-2-Dodecenoic acid; trans-dodec-2-enoic acid; 2E-Lauroleic acid; 2E-Dodecenoic acid; (E)-2-dodecenoic acid; 1289-45-8; C12:1n-10; TRANS-2-DODECENOICACID; 2-Dodecensaeure; 2t-Dodecensaeure; Dodec-2-ensaeure; Dodec-2-enoicacid; EINECS 224-569-1; Dodecen-(2t)-saeure; trans-2-lauroleic acid; 12:1, n-10 trans; C12:1, n-10 trans; CHEMBL4281333; CHEBI:37162; CHEBI:38371; DTXSID401343611; AMY37907; ZINC1689827; LMFA01030788; MFCD00045906; NSC 59856; AKOS008105134; AS-57983; WS-00320; AI3-11152; C12:1; CS-0152270; D5660; EN300-65552; EN300-365600; W16764; A826494; A934942; Q27117056; Q27117632; Z336127680; 3X1
|
|
| CAS | 1289-45-8 | |
| PubChem CID | 5282729 | |
| ChEMBL ID | CHEMBL4281333 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 198.3 | ALogp: | 4.8 |
| HBD: | 1 | HBA: | 2 |
| Rotatable Bonds: | 9 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 37.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 14 | QED Weighted: | 0.439 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.648 | MDCK Permeability: | 0.00002280 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.008 | 20% Bioavailability (F20%): | 0.008 |
| 30% Bioavailability (F30%): | 0.104 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.768 | Plasma Protein Binding (PPB): | 96.48% |
| Volume Distribution (VD): | 0.321 | Fu: | 3.13% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.073 | CYP1A2-substrate: | 0.211 |
| CYP2C19-inhibitor: | 0.029 | CYP2C19-substrate: | 0.282 |
| CYP2C9-inhibitor: | 0.205 | CYP2C9-substrate: | 0.977 |
| CYP2D6-inhibitor: | 0.003 | CYP2D6-substrate: | 0.104 |
| CYP3A4-inhibitor: | 0.018 | CYP3A4-substrate: | 0.033 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.313 | Half-life (T1/2): | 0.797 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.009 | Human Hepatotoxicity (H-HT): | 0.02 |
| Drug-inuced Liver Injury (DILI): | 0.021 | AMES Toxicity: | 0.006 |
| Rat Oral Acute Toxicity: | 0.026 | Maximum Recommended Daily Dose: | 0.019 |
| Skin Sensitization: | 0.871 | Carcinogencity: | 0.267 |
| Eye Corrosion: | 0.992 | Eye Irritation: | 0.992 |
| Respiratory Toxicity: | 0.289 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001587 | 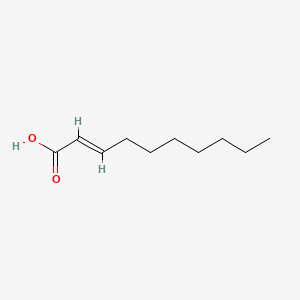 |
0.846 | D0Z5BC |  |
0.531 | ||
| ENC001590 |  |
0.684 | D0O1PH |  |
0.479 | ||
| ENC000088 |  |
0.636 | D0O1TC |  |
0.457 | ||
| ENC000267 |  |
0.628 | D0UE9X |  |
0.412 | ||
| ENC001656 |  |
0.617 | D05ATI | 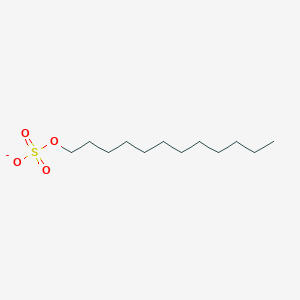 |
0.387 | ||
| ENC001655 | 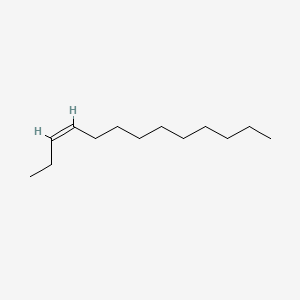 |
0.617 | D0XN8C |  |
0.380 | ||
| ENC001554 |  |
0.615 | D0N3NO |  |
0.378 | ||
| ENC001099 |  |
0.607 | D07ILQ |  |
0.375 | ||
| ENC001589 |  |
0.607 | D0E4WR |  |
0.370 | ||
| ENC000270 |  |
0.596 | D03ZJE |  |
0.361 | ||