NPs Basic Information
|
Name |
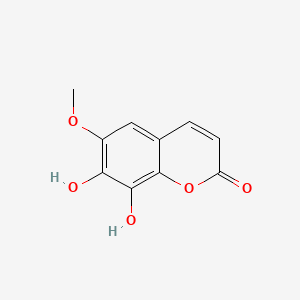
Fraxetin
|
| Molecular Formula | C10H8O5 | |
| IUPAC Name* |
7,8-dihydroxy-6-methoxychromen-2-one
|
|
| SMILES |
COC1=C(C(=C2C(=C1)C=CC(=O)O2)O)O
|
|
| InChI |
InChI=1S/C10H8O5/c1-14-6-4-5-2-3-7(11)15-10(5)9(13)8(6)12/h2-4,12-13H,1H3
|
|
| InChIKey |
HAVWRBANWNTOJX-UHFFFAOYSA-N
|
|
| Synonyms |
Fraxetin; 574-84-5; 7,8-Dihydroxy-6-methoxycoumarin; 7,8-Dihydroxy-6-methoxy-2H-chromen-2-one; 7,8-dihydroxy-6-methoxychromen-2-one; 2H-1-Benzopyran-2-one, 7,8-dihydroxy-6-methoxy-; 7,8-Dihydroxy-6-methoxy-2-benzopyrone; CD3GD44O3K; 7,8-Dihydroxy-6-methoxy-chromen-2-one; CHEMBL54909; CHEBI:5169; UNII-CD3GD44O3K; Fraxetol; 8-hydroxyscopoletin; EINECS 209-376-2; Spectrum_001507; SpecPlus_000477; FRAXETIN [MI]; Spectrum2_001639; Spectrum3_001842; Spectrum4_001686; Spectrum5_000332; 7,8-Dihydroxy-6-methoxy-2H-1-benzopyran-2-one; Oprea1_735469; SCHEMBL43472; BSPBio_003224; Fraxetin, analytical standard; KBioGR_001952; KBioSS_001987; MLS002207123; DivK1c_006573; SPECTRUM1504069; SPBio_001737; MEGxp0_000506; ACon0_001071; ACon1_000442; KBio1_001517; KBio2_001987; KBio2_004555; KBio2_007123; KBio3_002724; DTXSID00205992; 7,8-dihydroxy-6-methoxy coumarin; KUC106681N; ZINC113309; HY-N0580; TNP00177; Coumarin, 7,8-dihydroxy-6-methoxy; BDBM50206215; CCG-38759; MFCD00006873; s9503; STL564671; Coumarin, 7,8-dihydroxy-6-methoxy-; AKOS000277991; 7,8-Dihydroxy-6-methoxycoumarin, 98%; NCGC00017270-01; NCGC00017270-02; NCGC00017270-03; NCGC00017270-04; NCGC00017270-05; NCGC00096046-01; NCGC00096046-02; NCGC00169075-01; NCGC00169075-02; AC-34572; AS-67313; SMR000112323; KSC-11-207-12; DB-050316; CS-0009115; FT-0632418; 7,8-Dihydroxy-6-methoxy-2H-chromen-2-one #; A14554; C09265; 574F845; SR-05000002449; Q-100662; SR-05000002449-1; BRD-K76587808-001-03-8; Q15410973
|
|
| CAS | 574-84-5 | |
| PubChem CID | 5273569 | |
| ChEMBL ID | CHEMBL54909 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 208.17 | ALogp: | 1.2 |
| HBD: | 2 | HBA: | 5 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 76.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 15 | QED Weighted: | 0.55 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.922 | MDCK Permeability: | 0.00001540 |
| Pgp-inhibitor: | 0.006 | Pgp-substrate: | 0.032 |
| Human Intestinal Absorption (HIA): | 0.022 | 20% Bioavailability (F20%): | 0.039 |
| 30% Bioavailability (F30%): | 0.983 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.045 | Plasma Protein Binding (PPB): | 86.28% |
| Volume Distribution (VD): | 0.827 | Fu: | 15.29% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.9 | CYP1A2-substrate: | 0.944 |
| CYP2C19-inhibitor: | 0.032 | CYP2C19-substrate: | 0.067 |
| CYP2C9-inhibitor: | 0.107 | CYP2C9-substrate: | 0.512 |
| CYP2D6-inhibitor: | 0.418 | CYP2D6-substrate: | 0.409 |
| CYP3A4-inhibitor: | 0.114 | CYP3A4-substrate: | 0.141 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 15.221 | Half-life (T1/2): | 0.902 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.039 | Human Hepatotoxicity (H-HT): | 0.599 |
| Drug-inuced Liver Injury (DILI): | 0.955 | AMES Toxicity: | 0.117 |
| Rat Oral Acute Toxicity: | 0.244 | Maximum Recommended Daily Dose: | 0.194 |
| Skin Sensitization: | 0.807 | Carcinogencity: | 0.726 |
| Eye Corrosion: | 0.026 | Eye Irritation: | 0.594 |
| Respiratory Toxicity: | 0.262 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001623 | 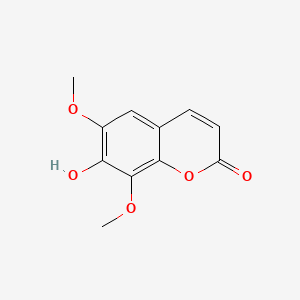 |
0.700 | D08SKH |  |
0.475 | ||
| ENC001537 |  |
0.569 | D0E9CD | 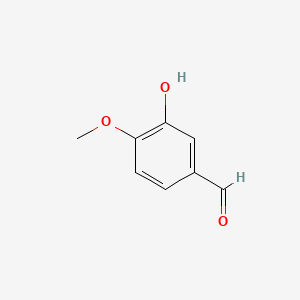 |
0.340 | ||
| ENC005232 |  |
0.460 | D06GCK | 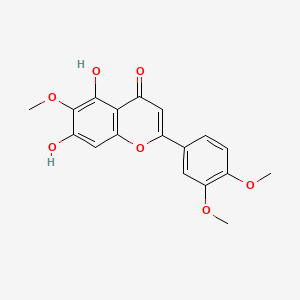 |
0.325 | ||
| ENC001561 | 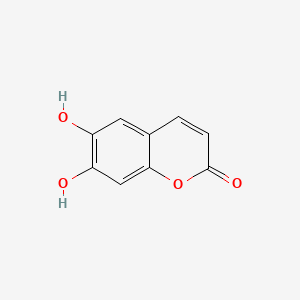 |
0.453 | D07MGA |  |
0.325 | ||
| ENC004401 |  |
0.441 | D0FA2O |  |
0.275 | ||
| ENC005717 |  |
0.439 | D0J4IX | 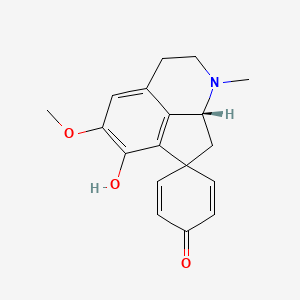 |
0.272 | ||
| ENC005716 |  |
0.439 | D0G4KG | 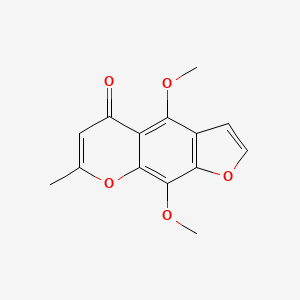 |
0.270 | ||
| ENC001472 | 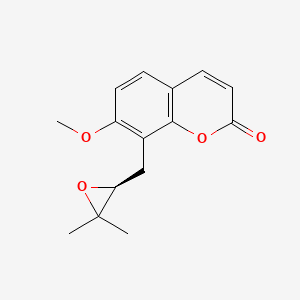 |
0.409 | D0U0OT | 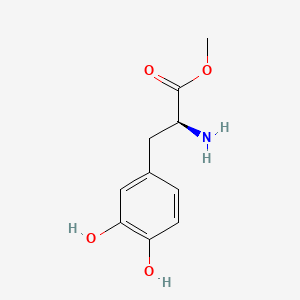 |
0.266 | ||
| ENC005905 |  |
0.403 | D0K8KX | 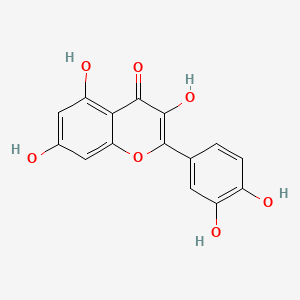 |
0.263 | ||
| ENC003710 | 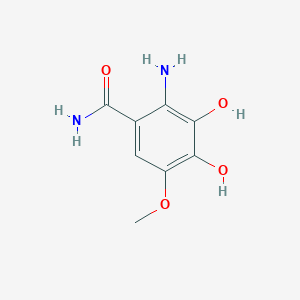 |
0.400 | D0DJ1B |  |
0.257 | ||