NPs Basic Information
|
Name |
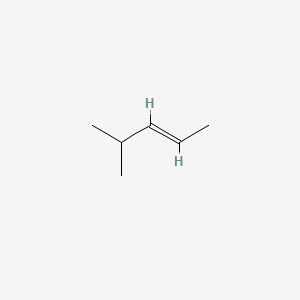
trans-4-Methyl-2-pentene
|
| Molecular Formula | C6H12 | |
| IUPAC Name* |
(E)-4-methylpent-2-ene
|
|
| SMILES |
C/C=C/C(C)C
|
|
| InChI |
InChI=1S/C6H12/c1-4-5-6(2)3/h4-6H,1-3H3/b5-4+
|
|
| InChIKey |
LGAQJENWWYGFSN-SNAWJCMRSA-N
|
|
| Synonyms |
trans-4-Methyl-2-pentene; 674-76-0; (E)-4-Methyl-2-pentene; (E)-4-Methylpent-2-ene; 4-METHYL-2-PENTENE; 2-Pentene, 4-methyl-; 2-Pentene, 4-methyl-, (E)-; 4-Methyl-trans-2-pentene; (2E)-4-methylpent-2-ene; 4-Methyl-2-pentene, (2E)-; 1,1-Dimethyl-2-butene; M38LU0DQ4J; (2E)-4-Methyl-2-pentene; NSC-73914; 2-Methyl-3-pentene; UNII-M38LU0DQ4J; MFCD00065138; EINECS 211-616-6; EINECS 224-721-7; trans-4-methylpentene-2; 4-Methyl-2-pentene,c&t; 4-Methyl-2-pentene(c,t); 4-Methylpentene-2, trans-; 4-Methyl-2-pentene, trans-; 4-Methyl-2-pentene, (E)-; (2E)-4-Methyl-2-pentene #; TRANS-2-METHYL-3-PENTENE; (E)-(CH3)2CHCH=CHCH3; DTXSID101015956; 2-Pentene, 4-methyl-, (2E)-; NSC73914; ZINC1699375; NSC 19873; NSC 73914; AKOS025295620; 2-PENTENE, 4-METHYL-, TRANS-; M0394; trans-4-Methyl-2-pentene, technical, >=90% (GC); Q27283426
|
|
| CAS | 674-76-0 | |
| PubChem CID | 172092 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 84.16 | ALogp: | 2.4 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 6 | QED Weighted: | 0.428 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.147 | MDCK Permeability: | 0.00002720 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.003 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.026 |
| 30% Bioavailability (F30%): | 0.57 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.981 | Plasma Protein Binding (PPB): | 96.39% |
| Volume Distribution (VD): | 3.195 | Fu: | 4.46% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.702 | CYP1A2-substrate: | 0.761 |
| CYP2C19-inhibitor: | 0.122 | CYP2C19-substrate: | 0.918 |
| CYP2C9-inhibitor: | 0.09 | CYP2C9-substrate: | 0.866 |
| CYP2D6-inhibitor: | 0.024 | CYP2D6-substrate: | 0.402 |
| CYP3A4-inhibitor: | 0.023 | CYP3A4-substrate: | 0.313 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 7.521 | Half-life (T1/2): | 0.579 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.007 | Human Hepatotoxicity (H-HT): | 0.06 |
| Drug-inuced Liver Injury (DILI): | 0.08 | AMES Toxicity: | 0.013 |
| Rat Oral Acute Toxicity: | 0.018 | Maximum Recommended Daily Dose: | 0.097 |
| Skin Sensitization: | 0.187 | Carcinogencity: | 0.056 |
| Eye Corrosion: | 0.98 | Eye Irritation: | 0.995 |
| Respiratory Toxicity: | 0.039 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001828 |  |
0.357 | D0M1PQ |  |
0.171 | ||
| ENC001703 |  |
0.316 | D03LGG |  |
0.169 | ||
| ENC002241 | 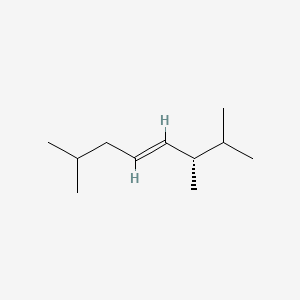 |
0.265 | D0U5CE | 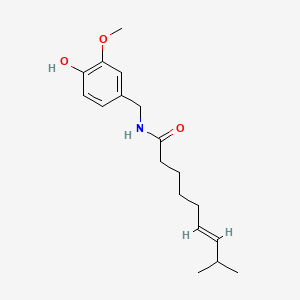 |
0.169 | ||
| ENC000619 |  |
0.250 | D0B2OT |  |
0.167 | ||
| ENC000147 |  |
0.238 | D0ZK8H |  |
0.167 | ||
| ENC000906 | 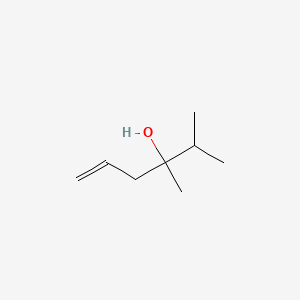 |
0.233 | D04CSZ |  |
0.158 | ||
| ENC000382 | 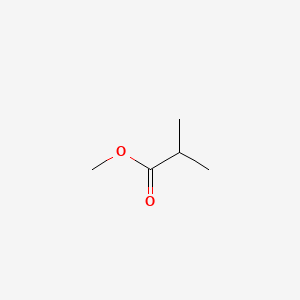 |
0.231 | D06GIP |  |
0.158 | ||
| ENC000237 | 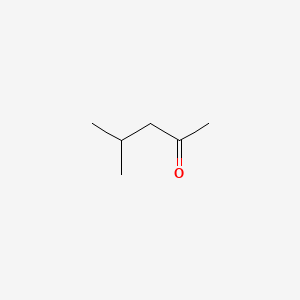 |
0.231 | D0T3NY | 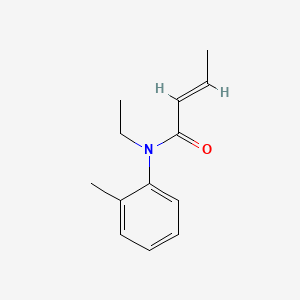 |
0.143 | ||
| ENC001734 | 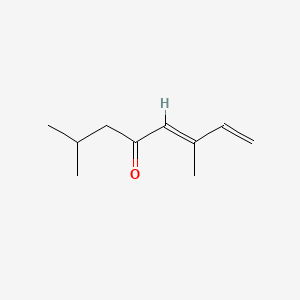 |
0.229 | D0A3HB |  |
0.140 | ||
| ENC001735 |  |
0.229 | D07ZTO | 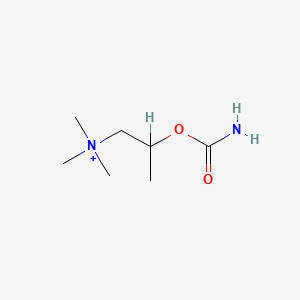 |
0.135 | ||