NPs Basic Information

|
Name |
Terreic acid
|
| Molecular Formula | C7H6O4 | |
| IUPAC Name* |
(1R,6S)-3-hydroxy-4-methyl-7-oxabicyclo[4.1.0]hept-3-ene-2,5-dione
|
|
| SMILES |
CC1=C(C(=O)[C@H]2[C@@H](C1=O)O2)O
|
|
| InChI |
InChI=1S/C7H6O4/c1-2-3(8)5(10)7-6(11-7)4(2)9/h6-8H,1H3/t6-,7+/m1/s1
|
|
| InChIKey |
ATFNSNUJZOYXFC-RQJHMYQMSA-N
|
|
| Synonyms |
Terreic acid; (-)-Terreic Acid; 121-40-4; (1R,6S)-3-Hydroxy-4-methyl-7-oxabicyclo[4.1.0]hept-3-ene-2,5-dione; XM2Y0DRJ7D; (1R)-3-Hydroxy-4-methyl-7-oxabicyclo(4.1.0)hept-3-ene-2,5-dione; NSC294734; NSC-294734; (1R,6S)-3-Hydroxy-4-methyl-7-oxabicyclo(4.1.0)hept-3-ene-2,5-dione; 5,6-Epoxy-3-hydroxy-p-toluquinone; UNII-XM2Y0DRJ7D; NSC 294734; (-)-TerreicAcid; 2-Hydroxy-3-methyl-1,4-benzoquinone 5,6-epoxide; BiomolKI_000073; BiomolKI2_000077; TERREIC ACID [MI]; 7-Oxabicyclo(4.1.0)hept-3-ene-2,5-dione, 3-hydroxy-4-methyl-, stereoisomer; SCHEMBL51229; CHEMBL1455300; DTXSID40879070; CHEBI:156546; HMS3267H06; HMS3412M22; HMS3676M22; 3-Hydroxy-4-methyl-7-oxabicyclo(4.1.0)hept-3-ene-2,5-dione; 7-Oxabicyclo(4.1.0)hept-3-ene-2,5-dione, 3-hydroxy-4-methyl-, (1R)-; AKOS006281919; ZINC100057173; CCG-100677; SMP2_000322; NCGC00025148-02; NCGC00025148-03; NCI60_002449; HY-110013; CS-0032828; (-)-Terreic Acid, Synthetic - CAS 121-40-4; (5R),(6S)-EPOXY-3-HYDROXY-P-TOLUQUINONE; SR-01000597558; SR-01000597558-1; Q27293905; 7-Oxabicyclo[4.1.0]hept-3-ene-2, 3-hydroxy-4-methyl-, (1R-cis)-; 7-Oxabicyclo[4.1.0]hept-3-ene-2,5-dione,3-hydroxy-4-methyl-,(1R,6S)-
|
|
| CAS | 121-40-4 | |
| PubChem CID | 91437 | |
| ChEMBL ID | CHEMBL1455300 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.12 | ALogp: | -0.2 |
| HBD: | 1 | HBA: | 4 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 66.9 | Aromatic Rings: | 2 |
| Heavy Atoms: | 11 | QED Weighted: | 0.505 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.076 | MDCK Permeability: | 0.00001660 |
| Pgp-inhibitor: | 0.008 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.012 | 20% Bioavailability (F20%): | 0.053 |
| 30% Bioavailability (F30%): | 0.001 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.058 | Plasma Protein Binding (PPB): | 89.40% |
| Volume Distribution (VD): | 0.458 | Fu: | 10.14% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.353 | CYP1A2-substrate: | 0.45 |
| CYP2C19-inhibitor: | 0.025 | CYP2C19-substrate: | 0.203 |
| CYP2C9-inhibitor: | 0.057 | CYP2C9-substrate: | 0.523 |
| CYP2D6-inhibitor: | 0.053 | CYP2D6-substrate: | 0.23 |
| CYP3A4-inhibitor: | 0.032 | CYP3A4-substrate: | 0.171 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.137 | Half-life (T1/2): | 0.851 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.042 | Human Hepatotoxicity (H-HT): | 0.107 |
| Drug-inuced Liver Injury (DILI): | 0.161 | AMES Toxicity: | 0.027 |
| Rat Oral Acute Toxicity: | 0.941 | Maximum Recommended Daily Dose: | 0.747 |
| Skin Sensitization: | 0.956 | Carcinogencity: | 0.483 |
| Eye Corrosion: | 0.288 | Eye Irritation: | 0.943 |
| Respiratory Toxicity: | 0.383 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC003178 |  |
0.415 | D0K7LU | 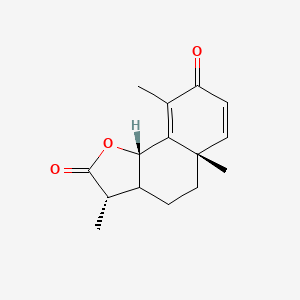 |
0.238 | ||
| ENC000951 | 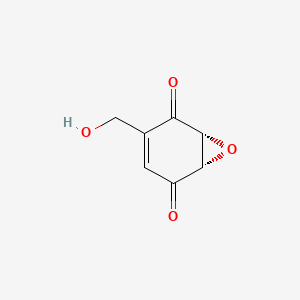 |
0.405 | D0Z8EX | 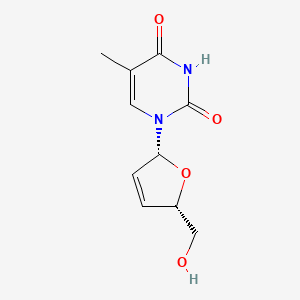 |
0.237 | ||
| ENC001362 |  |
0.311 | D0CL9S | 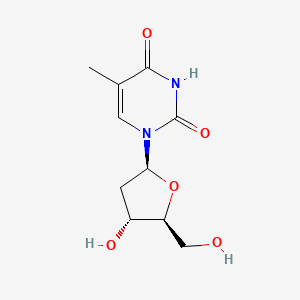 |
0.230 | ||
| ENC000868 |  |
0.310 | D03KXY |  |
0.213 | ||
| ENC004595 |  |
0.305 | D01XYJ |  |
0.209 | ||
| ENC004596 |  |
0.305 | D07AHW |  |
0.200 | ||
| ENC004597 |  |
0.305 | D0N0OU |  |
0.196 | ||
| ENC002293 |  |
0.304 | D0R2KF |  |
0.194 | ||
| ENC005552 |  |
0.295 | D0TS1Z |  |
0.190 | ||
| ENC000670 |  |
0.292 | D09PZO | 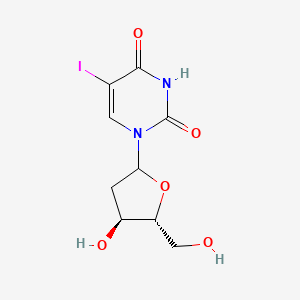 |
0.190 | ||