NPs Basic Information
|
Name |
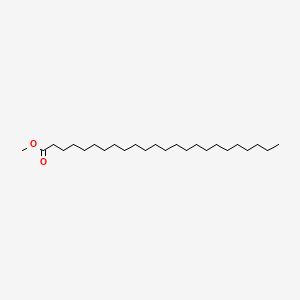
Methyl tetracosanoate
|
| Molecular Formula | C25H50O2 | |
| IUPAC Name* |
methyl tetracosanoate
|
|
| SMILES |
CCCCCCCCCCCCCCCCCCCCCCCC(=O)OC
|
|
| InChI |
InChI=1S/C25H50O2/c1-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20-21-22-23-24-25(26)27-2/h3-24H2,1-2H3
|
|
| InChIKey |
XUDJZDNUVZHSKZ-UHFFFAOYSA-N
|
|
| Synonyms |
Methyl tetracosanoate; 2442-49-1; Methyl lignocerate; Lignoceric acid methyl ester; Tetracosanoic acid, methyl ester; Methyl tetracosanoic acid; LIGNOCERICACIDMETHYLESTER; Tetracosanoic Acid Methyl Ester; lignoceric methyl ester; Tetracosanoic acid-methyl ester; HY5564B8FX; UNII-HY5564B8FX; Tetracosanoic acid,methyl ester; EINECS 219-475-2; MFCD00009351; Tetracosanoic acid methyl; SCHEMBL863540; DTXSID50179174; CDAA-251024M; CHEBI:143590; CAA44249; HY-N8438; ZINC70455481; AKOS015903349; AS-59413; Methyl tetracosanoate, analytical standard; DB-046446; Tetracosanoic acid methyl ester (FAME MIX); CS-0144211; FT-0634286; L0112; T72146; Methyl tetracosanoate, >=99.0% (capillary GC); J-015516; Q24762827; 2450B6FA-63F4-4631-86DE-6CDF98A1449A
|
|
| CAS | 2442-49-1 | |
| PubChem CID | 75546 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 382.7 | ALogp: | 12.3 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 23 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 27 | QED Weighted: | 0.124 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.103 | MDCK Permeability: | 0.00000670 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.88 |
| 30% Bioavailability (F30%): | 1 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.033 | Plasma Protein Binding (PPB): | 97.06% |
| Volume Distribution (VD): | 3.814 | Fu: | 1.10% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.08 | CYP1A2-substrate: | 0.161 |
| CYP2C19-inhibitor: | 0.18 | CYP2C19-substrate: | 0.056 |
| CYP2C9-inhibitor: | 0.057 | CYP2C9-substrate: | 0.966 |
| CYP2D6-inhibitor: | 0.211 | CYP2D6-substrate: | 0.024 |
| CYP3A4-inhibitor: | 0.229 | CYP3A4-substrate: | 0.025 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.59 | Half-life (T1/2): | 0.067 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.526 | Human Hepatotoxicity (H-HT): | 0.019 |
| Drug-inuced Liver Injury (DILI): | 0.505 | AMES Toxicity: | 0.006 |
| Rat Oral Acute Toxicity: | 0.012 | Maximum Recommended Daily Dose: | 0.016 |
| Skin Sensitization: | 0.972 | Carcinogencity: | 0.03 |
| Eye Corrosion: | 0.945 | Eye Irritation: | 0.911 |
| Respiratory Toxicity: | 0.81 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000553 |  |
0.929 | D00AOJ |  |
0.747 | ||
| ENC000464 |  |
0.923 | D07ILQ |  |
0.533 | ||
| ENC000474 |  |
0.846 | D00STJ |  |
0.512 | ||
| ENC000358 |  |
0.814 | D00FGR |  |
0.471 | ||
| ENC000497 |  |
0.808 | D0Z5SM |  |
0.435 | ||
| ENC000446 |  |
0.805 | D0O1PH |  |
0.412 | ||
| ENC000433 |  |
0.776 | D05ATI | 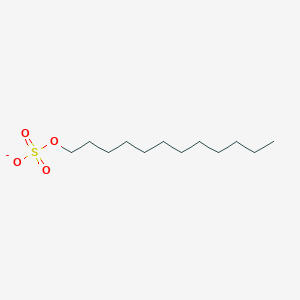 |
0.374 | ||
| ENC000282 | 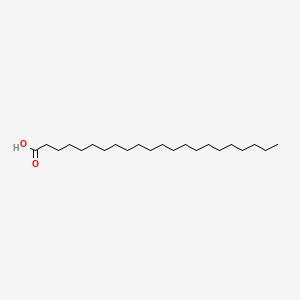 |
0.771 | D00MLW | 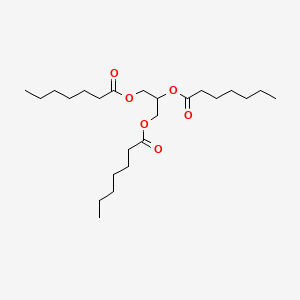 |
0.361 | ||
| ENC000280 |  |
0.769 | D0Z1QC | 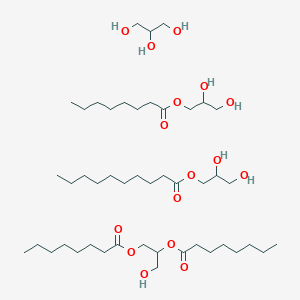 |
0.348 | ||
| ENC000442 |  |
0.768 | D0T9TJ |  |
0.344 | ||