NPs Basic Information
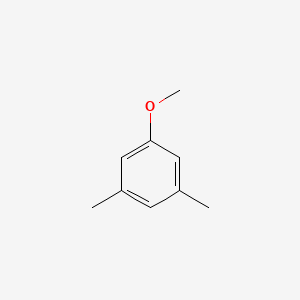
|
Name |
3,5-Dimethylanisole
|
| Molecular Formula | C9H12O | |
| IUPAC Name* |
1-methoxy-3,5-dimethylbenzene
|
|
| SMILES |
CC1=CC(=CC(=C1)OC)C
|
|
| InChI |
InChI=1S/C9H12O/c1-7-4-8(2)6-9(5-7)10-3/h4-6H,1-3H3
|
|
| InChIKey |
JCHJBEZBHANKGA-UHFFFAOYSA-N
|
|
| Synonyms |
3,5-Dimethylanisole; 874-63-5; 1-Methoxy-3,5-dimethylbenzene; Benzene, 1-methoxy-3,5-dimethyl-; 5-Methoxy-m-xylene; GP3L97PZB8; NSC-406910; 1-methoxy-3,5-dimethyl-benzene; 3,5-dimethylanisol; EINECS 212-865-3; MFCD00008398; NSC406910; 3,5-Dimethylanizole; AI3-11303; Anisole, 3,5-dimethyl; UNII-GP3L97PZB8; NCIOpen2_003733; SCHEMBL145855; 5-Methoxy-1,3-dimethylbenzene; SCHEMBL13322156; 3,5-Dimethylanisole, >=99%; 3,5-Dimethylphenyl methyl ether; JCHJBEZBHANKGA-UHFFFAOYSA-; DTXSID40236326; 1-Methoxy-3,5-dimethylbenzene #; ZINC967263; AMY12543; AKOS008948118; NSC 406910; PS-6237; DB-031939; CS-0146244; D2441; FT-0614686; EN300-86201; A842223; W-104021
|
|
| CAS | 874-63-5 | |
| PubChem CID | 70126 | |
| ChEMBL ID | NA |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 136.19 | ALogp: | 2.7 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 9.2 | Aromatic Rings: | 1 |
| Heavy Atoms: | 10 | QED Weighted: | 0.576 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.363 | MDCK Permeability: | 0.00002200 |
| Pgp-inhibitor: | 0.003 | Pgp-substrate: | 0.02 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.586 |
| 30% Bioavailability (F30%): | 0.7 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.955 | Plasma Protein Binding (PPB): | 91.16% |
| Volume Distribution (VD): | 1.299 | Fu: | 5.05% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.891 | CYP1A2-substrate: | 0.94 |
| CYP2C19-inhibitor: | 0.792 | CYP2C19-substrate: | 0.905 |
| CYP2C9-inhibitor: | 0.134 | CYP2C9-substrate: | 0.806 |
| CYP2D6-inhibitor: | 0.53 | CYP2D6-substrate: | 0.916 |
| CYP3A4-inhibitor: | 0.166 | CYP3A4-substrate: | 0.483 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 11.174 | Half-life (T1/2): | 0.75 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.047 | Human Hepatotoxicity (H-HT): | 0.184 |
| Drug-inuced Liver Injury (DILI): | 0.145 | AMES Toxicity: | 0.056 |
| Rat Oral Acute Toxicity: | 0.025 | Maximum Recommended Daily Dose: | 0.086 |
| Skin Sensitization: | 0.589 | Carcinogencity: | 0.184 |
| Eye Corrosion: | 0.976 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.113 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000736 | 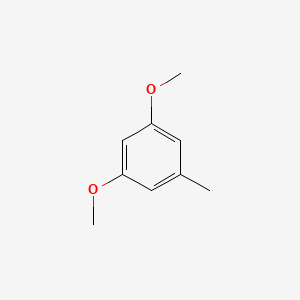 |
0.676 | D0S5CH |  |
0.440 | ||
| ENC000242 |  |
0.594 | D09GYT | 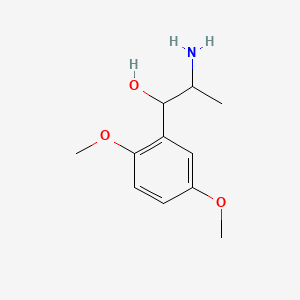 |
0.288 | ||
| ENC000491 |  |
0.440 | D06GIP |  |
0.273 | ||
| ENC001186 |  |
0.426 | D0DJ1B |  |
0.254 | ||
| ENC000349 | 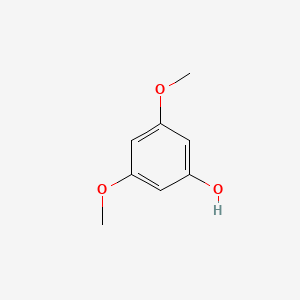 |
0.425 | D05CKR |  |
0.250 | ||
| ENC000979 |  |
0.411 | D0T1LK |  |
0.246 | ||
| ENC002445 |  |
0.396 | D0X0RI |  |
0.240 | ||
| ENC005289 |  |
0.393 | D0E9CD | 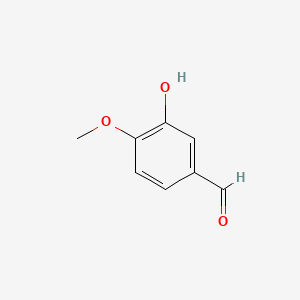 |
0.239 | ||
| ENC004151 | 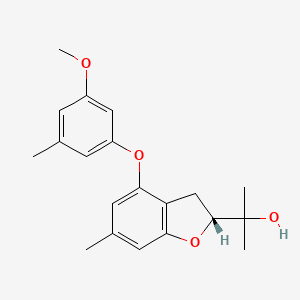 |
0.388 | D0FA2O |  |
0.233 | ||
| ENC005291 | 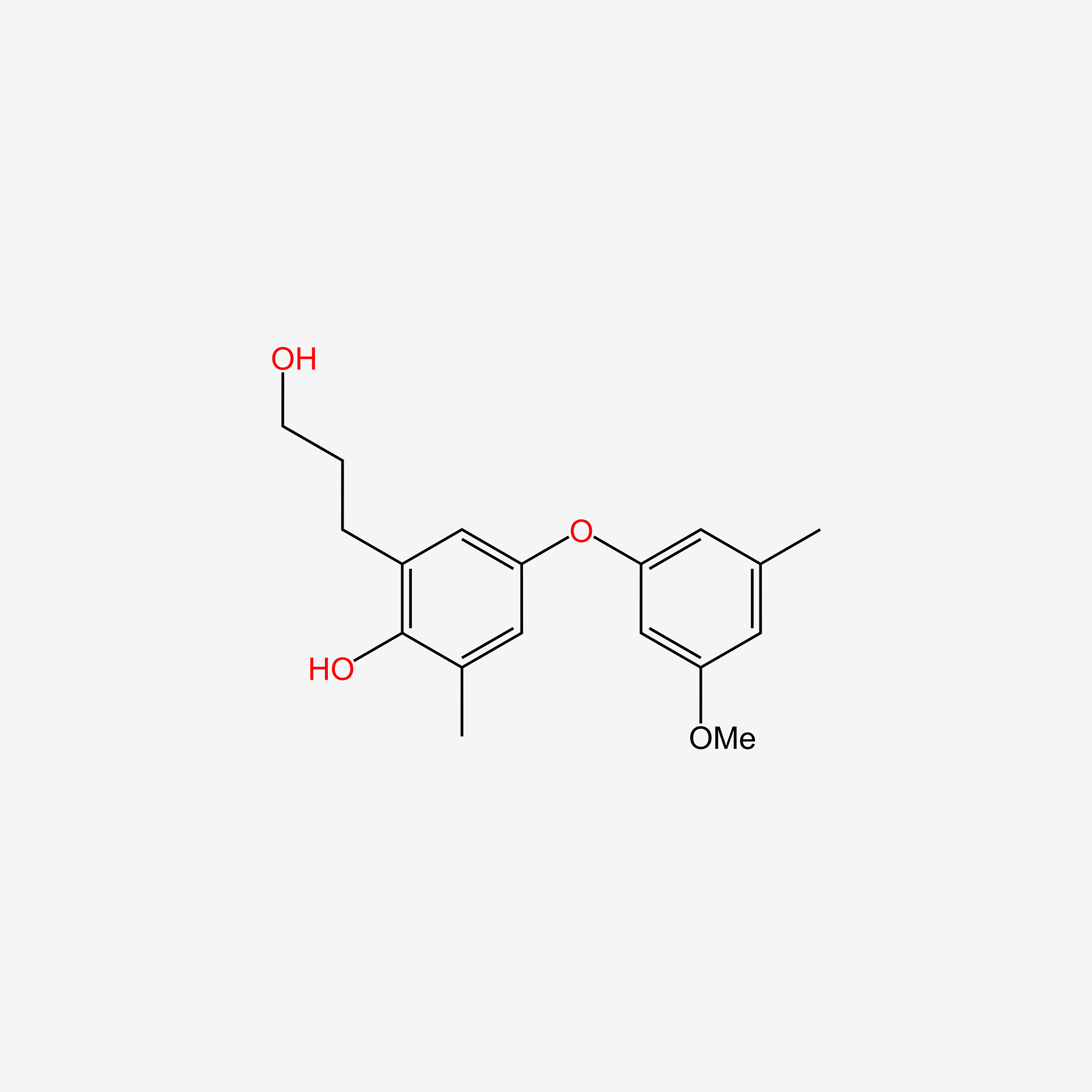 |
0.375 | D05VIX |  |
0.230 | ||