NPs Basic Information
|
Name |
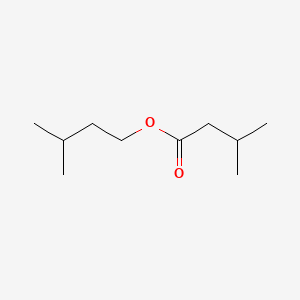
Isoamyl isovalerate
|
| Molecular Formula | C10H20O2 | |
| IUPAC Name* |
3-methylbutyl 3-methylbutanoate
|
|
| SMILES |
CC(C)CCOC(=O)CC(C)C
|
|
| InChI |
InChI=1S/C10H20O2/c1-8(2)5-6-12-10(11)7-9(3)4/h8-9H,5-7H2,1-4H3
|
|
| InChIKey |
XINCECQTMHSORG-UHFFFAOYSA-N
|
|
| Synonyms |
ISOAMYL ISOVALERATE; 659-70-1; Isopentyl 3-methylbutanoate; 3-Methylbutyl 3-methylbutanoate; Isopentyl isopentanoate; Isopentyl isovalerate; Solusterol; iso-Amyl isovalerate; 3-Methylbutyl isovalerate; Isoamyl valerianate; Apple essence; Butanoic acid, 3-methyl-, 3-methylbutyl ester; Isovaleric acid, isopentyl ester; Isoamyl 3-methylbutanoate; Isopentyl 3-methylbutyrate; Isoamyl 3-methylbutyrate; 3-Methylbutyl 3-methylbutyrate; Isoamyl isopentanoate; Isoamyl isovalerianate; Isopentyl alcohol, isovalerate; FEMA No. 2085; NSC 6565; Isovaleric Acid Isoamyl Ester; 16M1VA1FJY; 3-methylbutanoic acid 3-methylbutyl ester; CHEBI:31727; NSC-6565; Isoamyl isovalerate (natural); CCRIS 1348; EINECS 211-536-1; UNII-16M1VA1FJY; BRN 1753884; AI3-06045; iso-amyl iso-valerate; Isovaleric acid isoamyl; Nat.Isoamyl Isovalerate; ISOAMYLISOVALERATE; isoamyl 3-methyl butanoate; DSSTox_CID_24757; DSSTox_RID_80450; DSSTox_GSID_44757; Isopentyl 3-methylbutanoate #; SCHEMBL113763; Isovaleric Acid Isopentyl Ester; CHEMBL3189076; DTXSID8044757; ISOAMYL ISOVALERATE [MI]; FEMA 2085; XINCECQTMHSORG-UHFFFAOYSA-; ISOAMYL ISOVALERATE [FCC]; NSC6565; NATURAL ISOAMYL ISOVALERATE; ISOAMYL ISOVALERATE [FHFI]; ZINC1693629; Tox21_301620; WLN: 1Y1&2OV1Y1&1; LMFA07010919; MFCD00048369; AKOS015907822; (3-methyl-1-butyl) 3-methylbutanoate; Isoamyl isovalerate, >=98%, FCC, FG; NCGC00256163-01; AS-59218; CAS-659-70-1; 3-methyl-butyric acid 3-methyl-butyl ester; FT-0657233; I0196; D97798; Isoamyl isovalerate, natural, >=98%, FCC, FG; A835290; W-104778; Q11066025
|
|
| CAS | 659-70-1 | |
| PubChem CID | 12613 | |
| ChEMBL ID | CHEMBL3189076 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 172.26 | ALogp: | 3.1 |
| HBD: | 0 | HBA: | 2 |
| Rotatable Bonds: | 6 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 26.3 | Aromatic Rings: | 0 |
| Heavy Atoms: | 12 | QED Weighted: | 0.594 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.202 | MDCK Permeability: | 0.00003420 |
| Pgp-inhibitor: | 0.004 | Pgp-substrate: | 0.001 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.024 |
| 30% Bioavailability (F30%): | 0.348 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.811 | Plasma Protein Binding (PPB): | 81.25% |
| Volume Distribution (VD): | 0.807 | Fu: | 15.74% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.894 | CYP1A2-substrate: | 0.123 |
| CYP2C19-inhibitor: | 0.605 | CYP2C19-substrate: | 0.75 |
| CYP2C9-inhibitor: | 0.733 | CYP2C9-substrate: | 0.752 |
| CYP2D6-inhibitor: | 0.006 | CYP2D6-substrate: | 0.074 |
| CYP3A4-inhibitor: | 0.041 | CYP3A4-substrate: | 0.242 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 13.649 | Half-life (T1/2): | 0.708 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.016 | Human Hepatotoxicity (H-HT): | 0.05 |
| Drug-inuced Liver Injury (DILI): | 0.427 | AMES Toxicity: | 0.006 |
| Rat Oral Acute Toxicity: | 0.049 | Maximum Recommended Daily Dose: | 0.014 |
| Skin Sensitization: | 0.82 | Carcinogencity: | 0.328 |
| Eye Corrosion: | 0.973 | Eye Irritation: | 0.989 |
| Respiratory Toxicity: | 0.111 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000397 | 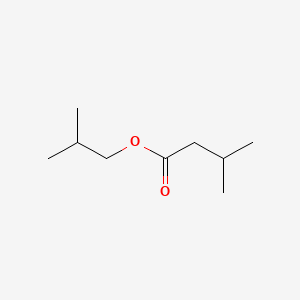 |
0.686 | D00WUF | 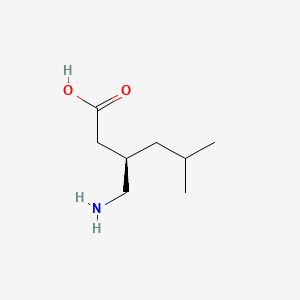 |
0.311 | ||
| ENC001137 | 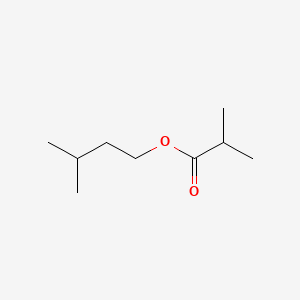 |
0.639 | D0Q9HF |  |
0.273 | ||
| ENC000227 | 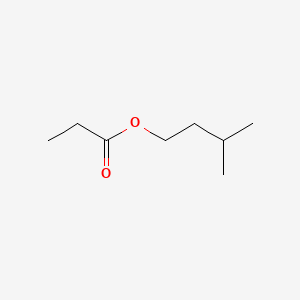 |
0.629 | D0B2OT |  |
0.261 | ||
| ENC000231 |  |
0.579 | D0K3LW |  |
0.246 | ||
| ENC000241 | 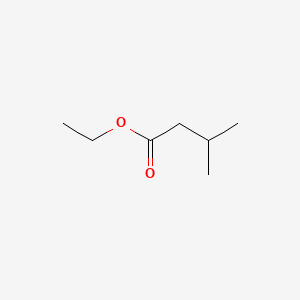 |
0.543 | D0U7BW | 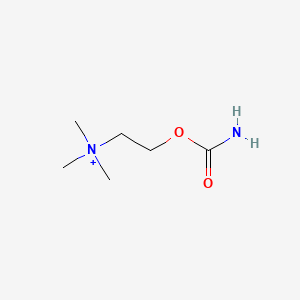 |
0.244 | ||
| ENC000603 |  |
0.543 | D0O6KE | 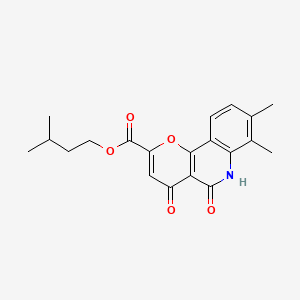 |
0.244 | ||
| ENC000718 |  |
0.537 | D0ZK8H |  |
0.244 | ||
| ENC000645 |  |
0.500 | D0P7VJ |  |
0.242 | ||
| ENC000187 |  |
0.436 | D05PLH |  |
0.238 | ||
| ENC001015 |  |
0.429 | D0I8CA |  |
0.236 | ||