NPs Basic Information

|
Name |
2,6-Dimethylnaphthalene
|
| Molecular Formula | C12H12 | |
| IUPAC Name* |
2,6-dimethylnaphthalene
|
|
| SMILES |
CC1=CC2=C(C=C1)C=C(C=C2)C
|
|
| InChI |
InChI=1S/C12H12/c1-9-3-5-12-8-10(2)4-6-11(12)7-9/h3-8H,1-2H3
|
|
| InChIKey |
YGYNBBAUIYTWBF-UHFFFAOYSA-N
|
|
| Synonyms |
2,6-DIMETHYLNAPHTHALENE; 581-42-0; Naphthalene, 2,6-dimethyl-; 2,6-DMN; 2,6-Dimethyl-naphthalene; 76U29QW3FM; CHEMBL194983; CHEBI:34251; C14330; MFCD00004120; NSC-36852; UNII-76U29QW3FM; EINECS 209-464-0; NSC 36852; Naphthalene,dimethyl-; Boronicacid, B-butyl-; Naphthalene, 2,6-(or 2,7)-dimethyl-; 2.6-dimethylnaphthalene; 2,6-dimethyl-naphtalene; AI3-01876; DSSTox_CID_9187; DSSTox_RID_78701; DSSTox_GSID_29187; BIDD:ER0559; 2,6-Dimethylnaphthalene, 99%; DTXSID0029187; AMY38494; NSC36852; ZINC1669605; Tox21_200260; BDBM50159257; STL507993; AKOS015842594; CS-W017996; NCGC00248581-01; NCGC00248581-02; NCGC00257814-01; 96789-56-9; CAS-581-42-0; J45.113D; LS-14057; DB-047460; A8250; D0751; FT-0625147; J-017269; Q-102015; Q2250787; F0001-1516; Dimethylnaphthalene (technical) 10 microg/mL in Cyclohexane
|
|
| CAS | 581-42-0 | |
| PubChem CID | 11387 | |
| ChEMBL ID | CHEMBL194983 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 156.22 | ALogp: | 4.3 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 2 |
| Heavy Atoms: | 12 | QED Weighted: | 0.536 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.43 | MDCK Permeability: | 0.00001710 |
| Pgp-inhibitor: | 0.095 | Pgp-substrate: | 0.017 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.498 |
| 30% Bioavailability (F30%): | 0.858 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.827 | Plasma Protein Binding (PPB): | 96.38% |
| Volume Distribution (VD): | 0.874 | Fu: | 3.69% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.947 | CYP1A2-substrate: | 0.917 |
| CYP2C19-inhibitor: | 0.683 | CYP2C19-substrate: | 0.559 |
| CYP2C9-inhibitor: | 0.421 | CYP2C9-substrate: | 0.751 |
| CYP2D6-inhibitor: | 0.594 | CYP2D6-substrate: | 0.92 |
| CYP3A4-inhibitor: | 0.26 | CYP3A4-substrate: | 0.466 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 10.436 | Half-life (T1/2): | 0.328 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.08 | Human Hepatotoxicity (H-HT): | 0.121 |
| Drug-inuced Liver Injury (DILI): | 0.598 | AMES Toxicity: | 0.541 |
| Rat Oral Acute Toxicity: | 0.055 | Maximum Recommended Daily Dose: | 0.667 |
| Skin Sensitization: | 0.724 | Carcinogencity: | 0.807 |
| Eye Corrosion: | 0.832 | Eye Irritation: | 0.994 |
| Respiratory Toxicity: | 0.036 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000169 |  |
0.571 | D0DJ1B |  |
0.421 | ||
| ENC000233 |  |
0.474 | D05CKR |  |
0.414 | ||
| ENC000239 |  |
0.436 | D02NTO |  |
0.347 | ||
| ENC000180 |  |
0.381 | D08GSF |  |
0.333 | ||
| ENC000221 |  |
0.372 | D0Y7PG |  |
0.313 | ||
| ENC000086 |  |
0.366 | D06GIP |  |
0.313 | ||
| ENC001367 | 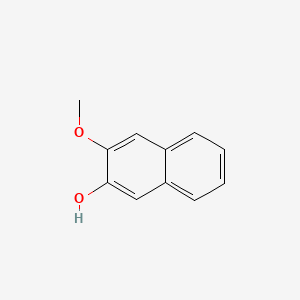 |
0.365 | D09BHB |  |
0.303 | ||
| ENC000498 |  |
0.356 | D03WEX |  |
0.301 | ||
| ENC000199 | 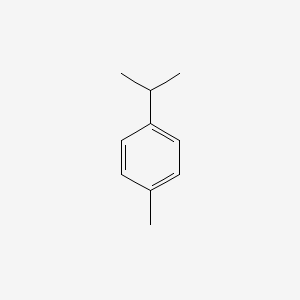 |
0.356 | D02WCI |  |
0.298 | ||
| ENC000552 |  |
0.356 | D0J6WW | 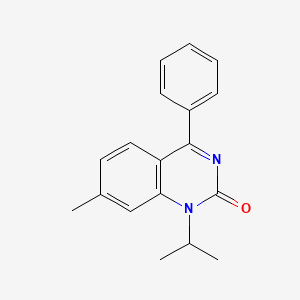 |
0.292 | ||