NPs Basic Information

|
Name |
2-Methylindole
|
| Molecular Formula | C9H9N | |
| IUPAC Name* |
2-methyl-1H-indole
|
|
| SMILES |
CC1=CC2=CC=CC=C2N1
|
|
| InChI |
InChI=1S/C9H9N/c1-7-6-8-4-2-3-5-9(8)10-7/h2-6,10H,1H3
|
|
| InChIKey |
BHNHHSOHWZKFOX-UHFFFAOYSA-N
|
|
| Synonyms |
2-METHYLINDOLE; 2-Methyl-1H-indole; 95-20-5; 1H-Indole, 2-methyl-; Indole, 2-methyl-; 2-METHYL INDOLE; CHEBI:49402; I7CN58827I; NSC-7514; 2-methylindol; NSC 7514; EINECS 202-398-3; MFCD00005616; Methylketole; methyl indole; UNII-I7CN58827I; AI3-03945; 2-methyl-indole; 2-methyl-1h-indol; 2-methyl-1-H-indole; 2-Methylindole, 98%; SCHEMBL12420; SCHEMBL377807; CHEMBL259419; F0290-0682; DTXSID5059117; NSC7514; KUC106612N; 2-Methylindole, analytical standard; ACT07545; AMY23231; KSC-09-215A; STR01200; ZINC1088076; AC-611; STK044221; AKOS000119568; CG-0500; CS-W007556; SB14955; NCGC00342152-01; DB-031753; 2-Methylindole, purum, >=97.5% (HPLC); FT-0612877; FT-0613057; M0346; EN300-18136; M-3895; AB00375786-04; Q4596907; 2MI
|
|
| CAS | 95-20-5 | |
| PubChem CID | 7224 | |
| ChEMBL ID | CHEMBL259419 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 131.17 | ALogp: | 2.5 |
| HBD: | 1 | HBA: | 0 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 15.8 | Aromatic Rings: | 2 |
| Heavy Atoms: | 10 | QED Weighted: | 0.565 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.494 | MDCK Permeability: | 0.00001970 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.033 |
| Human Intestinal Absorption (HIA): | 0.004 | 20% Bioavailability (F20%): | 0.298 |
| 30% Bioavailability (F30%): | 0.074 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.846 | Plasma Protein Binding (PPB): | 87.40% |
| Volume Distribution (VD): | 1.469 | Fu: | 10.74% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.973 | CYP1A2-substrate: | 0.943 |
| CYP2C19-inhibitor: | 0.668 | CYP2C19-substrate: | 0.183 |
| CYP2C9-inhibitor: | 0.106 | CYP2C9-substrate: | 0.653 |
| CYP2D6-inhibitor: | 0.768 | CYP2D6-substrate: | 0.897 |
| CYP3A4-inhibitor: | 0.126 | CYP3A4-substrate: | 0.248 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 8.237 | Half-life (T1/2): | 0.704 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.031 | Human Hepatotoxicity (H-HT): | 0.325 |
| Drug-inuced Liver Injury (DILI): | 0.274 | AMES Toxicity: | 0.801 |
| Rat Oral Acute Toxicity: | 0.675 | Maximum Recommended Daily Dose: | 0.785 |
| Skin Sensitization: | 0.71 | Carcinogencity: | 0.661 |
| Eye Corrosion: | 0.669 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.979 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000041 | 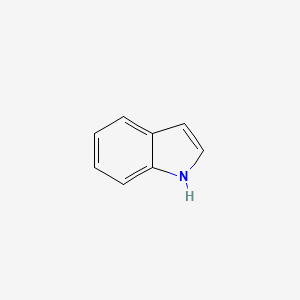 |
0.474 | D01PZD |  |
0.356 | ||
| ENC001388 | 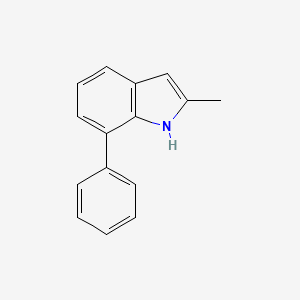 |
0.462 | D08QCJ |  |
0.340 | ||
| ENC000169 | 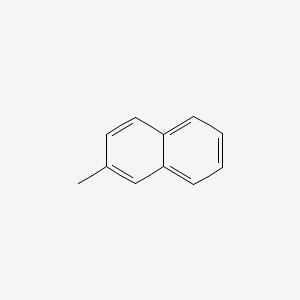 |
0.452 | D0K1XK |  |
0.333 | ||
| ENC000167 | 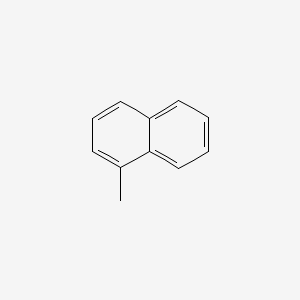 |
0.419 | D09JUG | 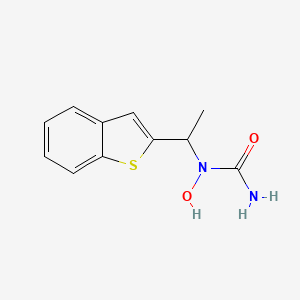 |
0.327 | ||
| ENC000064 | 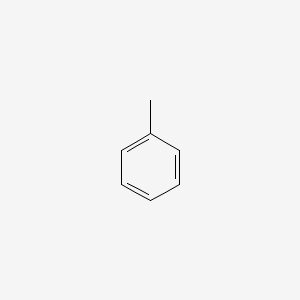 |
0.400 | D03GET |  |
0.327 | ||
| ENC000341 | 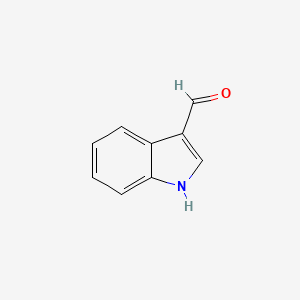 |
0.386 | D06DLI |  |
0.320 | ||
| ENC000179 |  |
0.378 | D05EJG | 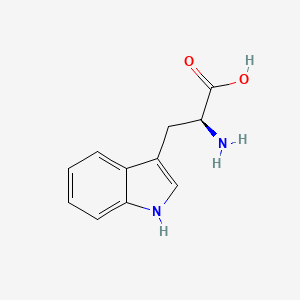 |
0.315 | ||
| ENC000028 |  |
0.378 | D02WCI |  |
0.302 | ||
| ENC001345 | 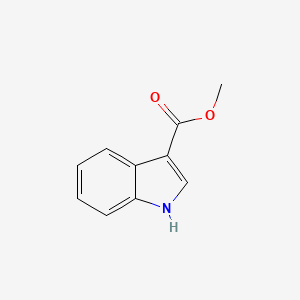 |
0.375 | D05OIS |  |
0.300 | ||
| ENC001367 | 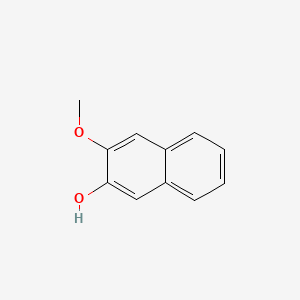 |
0.375 | D0T3LF |  |
0.295 | ||