NPs Basic Information
|
Name |
3-Hydroxybutyric acid
|
| Molecular Formula | C4H8O3 | |
| IUPAC Name* |
3-hydroxybutanoic acid
|
|
| SMILES |
CC(CC(=O)O)O
|
|
| InChI |
InChI=1S/C4H8O3/c1-3(5)2-4(6)7/h3,5H,2H2,1H3,(H,6,7)
|
|
| InChIKey |
WHBMMWSBFZVSSR-UHFFFAOYSA-N
|
|
| Synonyms |
3-hydroxybutyric acid; 3-hydroxybutanoic acid; 300-85-6; Butanoic acid, 3-hydroxy-; beta-Hydroxybutyric acid; 625-71-8; DL-beta-Hydroxybutyric acid; 3 HBA; DL-3-Hydroxybutyric Acid; 3-Hydroxybuttersaeure; Butyric acid, 3-hydroxy-; beta-Hydroxybuttersaeure; beta-hydroxybutyrate; beta-Hydroxy-n-butyric acid; 3-hydroxy-butanoic acid; .beta.-Hydroxybutyric acid; (1)-3-Hydroxybutyric acid; (+-)-3-Hydroxybutyric acid; beta-hydroxy-butyrate; (+/-)-3-Hydroxybutyric Acid; D,l-3 hydroxybutyrate; beta-hydroxybutanoic acid; NSC 3806; NSC-3806; Hydroxybutyric acid, dl-; .beta.-Hydroxy-n-butyric acid; AI3-21675; 26063-00-3; D(-)-beta-hydroxy butyric acid; CHEBI:20067; 3-hydroxybutyric acid, (+/-)-; TZP1275679; Butyric acid, 3-hydroxy- (8CI); SMR000112209; 3-HYDROXYBUTYRICACID; UNII-TZP1275679; EINECS 206-099-9; EINECS 210-908-0; 3-OH-butyric acid; BHBA; 3- hydroxybutyric acid; 3-hydroxy-butyric acid; DL-3-HydroxybutyricAcid; bmse000161; bmse000905; SCHEMBL2731; (RS)-3-hydroxybutyric acid; (+/-)-beta-Hydroxybutyrate; MLS001332397; MLS001332398; 3-Hydroxybutyric acid, 95%; DL-.beta.-Hydroxybutyric acid; (+/-)-3-Hydroxybutanoic acid; CHEMBL1162496; .BETA.HYDROXYBUTYRIC ACID; DTXSID60859511; NSC3806; (.+/-.)-3-Hydroxybutyric acid; HMS2270C20; HMS3369F13; AMY10050; BBL027467; LMFA01050005; MFCD00004546; s1031; STL377875; 3-HYDROXYBUTYRIC ACID [INCI]; AKOS009156821; AB88579; AC-5701; SB44475; .BETA.-HYDROXYBUTYRIC ACID [MI]; BETA-HYDROXYBUTYRIC ACID [WHO-DD]; VS-08544; HY-113378; CS-0062335; FT-0605155; FT-0605325; FT-0615836; FT-0625402; H0228; D84191; EN300-147463; A833855; A876308; Q223092; J-016241; J-017776; 869DD3C9-3114-4BE2-B4A6-6F1787476E95
|
|
| CAS | 300-85-6 | |
| PubChem CID | 441 | |
| ChEMBL ID | CHEMBL1162496 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 104.1 | ALogp: | -0.5 |
| HBD: | 2 | HBA: | 3 |
| Rotatable Bonds: | 2 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 57.5 | Aromatic Rings: | 0 |
| Heavy Atoms: | 7 | QED Weighted: | 0.528 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.45 | MDCK Permeability: | 0.00371931 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.012 |
| Human Intestinal Absorption (HIA): | 0.017 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.795 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.917 | Plasma Protein Binding (PPB): | 9.54% |
| Volume Distribution (VD): | 0.25 | Fu: | 78.78% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.013 | CYP1A2-substrate: | 0.087 |
| CYP2C19-inhibitor: | 0.026 | CYP2C19-substrate: | 0.09 |
| CYP2C9-inhibitor: | 0.004 | CYP2C9-substrate: | 0.913 |
| CYP2D6-inhibitor: | 0.012 | CYP2D6-substrate: | 0.193 |
| CYP3A4-inhibitor: | 0.008 | CYP3A4-substrate: | 0.047 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 6.902 | Half-life (T1/2): | 0.809 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.008 | Human Hepatotoxicity (H-HT): | 0.164 |
| Drug-inuced Liver Injury (DILI): | 0.047 | AMES Toxicity: | 0.019 |
| Rat Oral Acute Toxicity: | 0.026 | Maximum Recommended Daily Dose: | 0.025 |
| Skin Sensitization: | 0.165 | Carcinogencity: | 0.044 |
| Eye Corrosion: | 0.955 | Eye Irritation: | 0.991 |
| Respiratory Toxicity: | 0.036 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
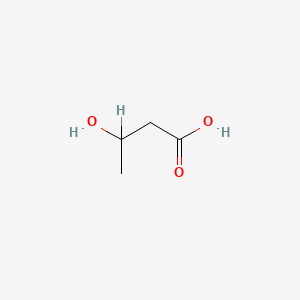
| ENC000351 | 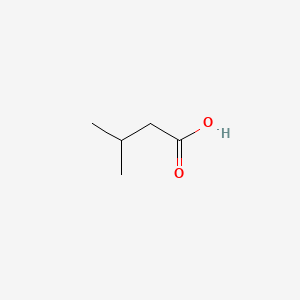 |
0.545 | D08QGD | 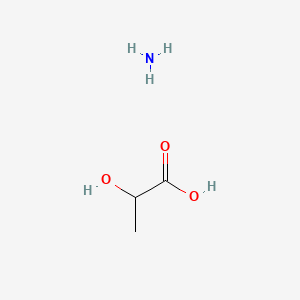 |
0.476 | ||
| ENC000037 | 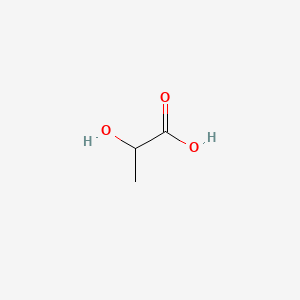 |
0.500 | D00WUF | 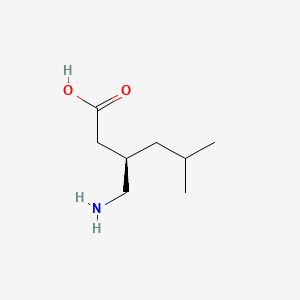 |
0.364 | ||
| ENC000057 |  |
0.400 | D09PUL | 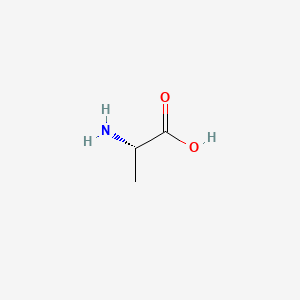 |
0.364 | ||
| ENC000058 |  |
0.400 | D00ZOF | 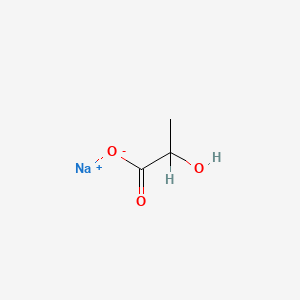 |
0.348 | ||
| ENC000824 | 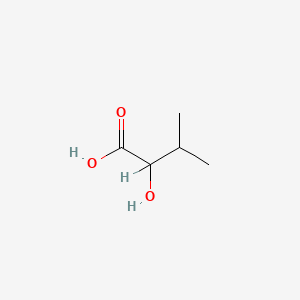 |
0.385 | D0M8AB |  |
0.333 | ||
| ENC000445 |  |
0.370 | D04CRL |  |
0.316 | ||
| ENC000010 |  |
0.364 | D0A8CJ |  |
0.313 | ||
| ENC000149 | 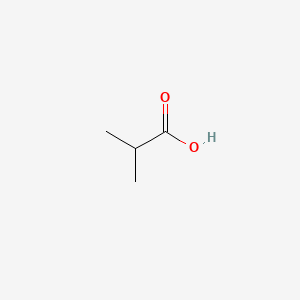 |
0.364 | D02UDJ | 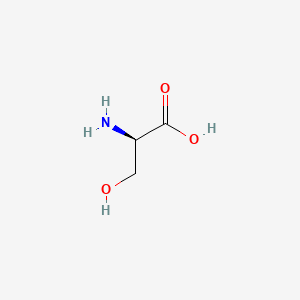 |
0.308 | ||
| ENC000890 | 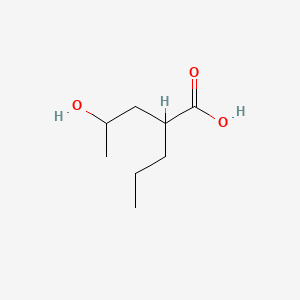 |
0.364 | D0EP8X |  |
0.296 | ||
| ENC000289 | 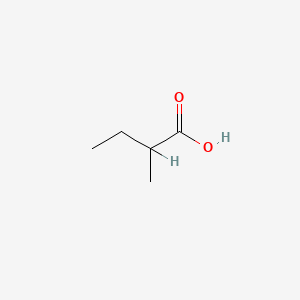 |
0.360 | D02KJX |  |
0.276 | ||