NPs Basic Information
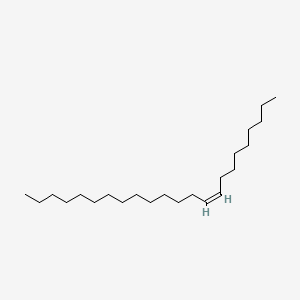
|
Name |
(Z)-9-Tricosene
|
| Molecular Formula | C23H46 | |
| IUPAC Name* |
(Z)-tricos-9-ene
|
|
| SMILES |
CCCCCCCCCCCCC/C=C\CCCCCCCC
|
|
| InChI |
InChI=1S/C23H46/c1-3-5-7-9-11-13-15-17-19-21-23-22-20-18-16-14-12-10-8-6-4-2/h17,19H,3-16,18,20-23H2,1-2H3/b19-17-
|
|
| InChIKey |
IGOWHGRNPLFNDJ-ZPHPHTNESA-N
|
|
| Synonyms |
Muscalure; cis-9-Tricosene; 27519-02-4; (Z)-9-Tricosene; (Z)-tricos-9-ene; 9-Tricosene, (9Z)-; cis-Tricos-9-ene; 9-Tricosene, (Z)-; 9Z-Tricosene; (9Z)-Tricosene; 6BSP6HFW73; Flybait; Muscamone; Caswell No. 883C; (9Z)-9-Tricosene; EINECS 248-505-7; ENT 35349; UNII-6BSP6HFW73; EPA Pesticide Chemical Code 103201; BRN 1841622; AI3-35349; MFCD00008988; (9Z)-tricos-9-ene; MUSCALURE [MI]; Z-9-TRICOSENE; (9Z)-9-Tricosene #; TRICOSENE, Z-9-; (Z)-9-Tricosene, 97%; CHEMBL1894305; DTXSID0032653; ZINC6920413; LMFA11000117; 9Z-Tricosene CAS 27519-02-4; AKOS024462408; AM84868; NCGC00163763-01; AS-14813; T1242; Muscalure, PESTANAL(R), analytical standard; A819098; Q135445; J-016797
|
|
| CAS | 27519-02-4 | |
| PubChem CID | 5365075 | |
| ChEMBL ID | CHEMBL1894305 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 322.6 | ALogp: | 11.7 |
| HBD: | 0 | HBA: | 0 |
| Rotatable Bonds: | 19 | Lipinski's rule of five: | Rejected |
| Polar Surface Area: | 0.0 | Aromatic Rings: | 0 |
| Heavy Atoms: | 23 | QED Weighted: | 0.145 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.032 | MDCK Permeability: | 0.00001020 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.011 | 20% Bioavailability (F20%): | 0.659 |
| 30% Bioavailability (F30%): | 0.999 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.021 | Plasma Protein Binding (PPB): | 100.05% |
| Volume Distribution (VD): | 5.206 | Fu: | 0.87% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.08 | CYP1A2-substrate: | 0.169 |
| CYP2C19-inhibitor: | 0.206 | CYP2C19-substrate: | 0.061 |
| CYP2C9-inhibitor: | 0.057 | CYP2C9-substrate: | 0.968 |
| CYP2D6-inhibitor: | 0.321 | CYP2D6-substrate: | 0.131 |
| CYP3A4-inhibitor: | 0.226 | CYP3A4-substrate: | 0.035 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 4.053 | Half-life (T1/2): | 0.165 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.591 | Human Hepatotoxicity (H-HT): | 0.012 |
| Drug-inuced Liver Injury (DILI): | 0.079 | AMES Toxicity: | 0.023 |
| Rat Oral Acute Toxicity: | 0.026 | Maximum Recommended Daily Dose: | 0.051 |
| Skin Sensitization: | 0.973 | Carcinogencity: | 0.029 |
| Eye Corrosion: | 0.99 | Eye Irritation: | 0.967 |
| Respiratory Toxicity: | 0.425 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC001689 | 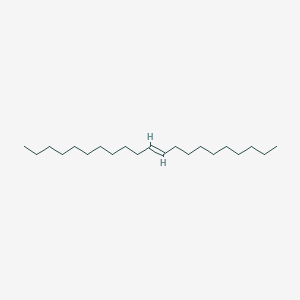 |
0.910 | D00AOJ |  |
0.595 | ||
| ENC001674 | 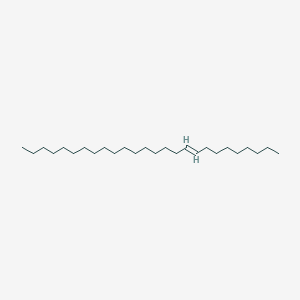 |
0.882 | D0O1PH |  |
0.583 | ||
| ENC001706 | 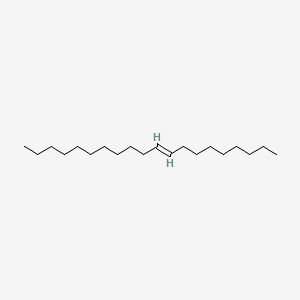 |
0.866 | D07ILQ |  |
0.512 | ||
| ENC001692 |  |
0.838 | D00FGR |  |
0.495 | ||
| ENC001681 |  |
0.821 | D0Z5SM |  |
0.458 | ||
| ENC001707 |  |
0.786 | D0O1TC |  |
0.398 | ||
| ENC000428 |  |
0.768 | D05ATI |  |
0.390 | ||
| ENC001710 |  |
0.766 | D0T9TJ |  |
0.387 | ||
| ENC001553 |  |
0.766 | D00STJ |  |
0.382 | ||
| ENC000285 |  |
0.761 | D0OR6A |  |
0.370 | ||