NPs Basic Information
|
Name |
Ellagic Acid
|
| Molecular Formula | C14H6O8 | |
| IUPAC Name* |
6,7,13,14-tetrahydroxy-2,9-dioxatetracyclo[6.6.2.04,16.011,15]hexadeca-1(15),4,6,8(16),11,13-hexaene-3,10-dione
|
|
| SMILES |
C1=C2C3=C(C(=C1O)O)OC(=O)C4=CC(=C(C(=C43)OC2=O)O)O
|
|
| InChI |
InChI=1S/C14H6O8/c15-5-1-3-7-8-4(14(20)22-11(7)9(5)17)2-6(16)10(18)12(8)21-13(3)19/h1-2,15-18H
|
|
| InChIKey |
AFSDNFLWKVMVRB-UHFFFAOYSA-N
|
|
| Synonyms |
ellagic acid; 476-66-4; Benzoaric acid; Lagistase; Elagostasine; 2,3,7,8-Tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione; Eleagic acid; Alizarine Yellow; Gallogen; Llagic acid; Ellagicacid; Acide ellagique; Acido elagico; Acidum ellagicum; C.I. 55005; C.I. 75270; Ellagate; Gallogen, astringent; MLS000069632; NSC407286; NSC-407286; NSC-656272; 19YRN3ZS9P; CHEMBL6246; SMR000058244; CHEBI:4775; 4,4',5,5',6,6'-hexahydroxydiphenic acid 2,6,2',6'-dilactone; 2,3,7,8-Tetrahydroxy(1)benzopyrano(5,4,3-cde)(1)benzopyran-5,10-dione; NSC656272; tetrahydroxy[?]dione; NCGC00017245-02; DSSTox_CID_557; (1)Benzopyrano(5,4,3-cde)(1)benzopyran-5,10-dione, 2,3,7,8-tetrahydroxy-; 2,3,7,8-Tetrahydroxy(1)benzopyrano(5,4,3-cde)-(1)benzopyran-5,10-dione; 2,3,7,8-tetrahydroxy[1]benzopyrano[5,4,3-cde][1]benzopyran-5,10-dione; DSSTox_RID_75657; DSSTox_GSID_20557; 6,7,13,14-tetrahydroxy-2,9-dioxatetracyclo[6.6.2.04,16.011,15]hexadeca-1(15),4,6,8(16),11,13-hexaene-3,10-dione; Gallogen (VAN); Gallogen (astringent); Pomegranate juice; [1]Benzopyrano[5,4,3-cde][1]benzopyran-5,10-dione, 2,3,7,8-tetrahydroxy-; Ellagic acid [INN:DCF]; CAS-476-66-4; Acido elagico [INN-Spanish]; CCRIS 774; Acide ellagique [INN-French]; Acidum ellagicum [INN-Latin]; SR-01000721925; EINECS 207-508-3; MFCD00006914; UNII-19YRN3ZS9P; NSC 407286; NSC 656272; BRN 0047549; Benzoarate; Ellagsaeure; Eleagate; Ellagic; Llagate; ellagic-acid; Elagic Acid; HSDB 7574; 2zjw; REF; Ellagic acid, 96%; Spectrum_001194; Diphenic acid, 4,4',5,5',6,6'-hexahydroxy-, di-delta-lactone; Spectrum2_000905; Spectrum3_001535; Spectrum4_000750; Spectrum5_000959; ELLAGIC ACID [MI]; ELLAGIC ACID [INN]; ELLAGIC ACID [HSDB]; ELLAGIC ACID [INCI]; Oprea1_032884; SCHEMBL20429; BSPBio_002950; KBioGR_001080; KBioSS_001674; 5-19-07-00108 (Beilstein Handbook Reference); MLS006011868; BIDD:ER0482; BIDD:GT0565; SPECTRUM1502245; SPBio_000750; ELLAGIC ACID [WHO-DD]; BDBM4078; LTK-20; DTXSID2020557; SCHEMBL19184504; BCBcMAP01_000154; cid_5281855; KBio2_001674; KBio2_004242; KBio2_006810; KBio3_002450; Ellagic acid, analytical standard; HMS1921N20; HMS3673M09; Pharmakon1600-01502245; BCP10830; HY-B0183; TNP00132; ZINC3872446; Tox21_110805; BBL009292; CCG-36358; NSC758198; s1327; STK801964; Ellagic acid, >=96.0% (HPCE); AKOS004120045; Tox21_110805_1; CS-2067; DB08846; NSC-758198; SDCCGMLS-0066664.P001; SMP1_000111; NCGC00017245-01; NCGC00017245-03; NCGC00017245-04; NCGC00017245-05; NCGC00017245-06; NCGC00017245-07; NCGC00017245-08; NCGC00017245-09; NCGC00017245-10; NCGC00017245-12; NCGC00094975-01; NCGC00094975-02; NCGC00094975-03; NCGC00094975-04; NCGC00178375-01; AC-11647; AC-30710; AS-35095; NCI60_003869; SBI-0051742.P002; E0375; Ellagic Acid, Dihydrate - CAS 476-66-4; FT-0603405; MLS-0066664.0001; AB00052292_11; AB00052292_12; EN300-6472707; A872094; Q422044; SR-01000721925-3; SR-01000721925-4; SR-01000721925-5; SR-01000721925-6; SR-01000721925-7; W-202834; BRD-K30466858-001-05-4; Ellagic acid, primary pharmaceutical reference standard; Diphenic acid,4',5,5',6,6'-hexahydroxy-, di-.delta.-lactone; 2,3,7,8-Tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione #; Diphenic acid, 4,4',5,5',6,6'-hexahydroxy-, di-.delta.-lactone; [1]Benzopyrano[5,3-cde][1]benzopyran-5,10-dione, 2,3,7,8-tetrahydroxy-; 2,3,7,8-Tetrahydroxy-[1]benzopyrano[5,4,3-cde][1]benzopyran-5,10-dione; 2,3,7,8-TETRAHYDROXY-CHROMENO[5,4,3-CDE]CHROMENE-5,10-DIONE; 2,3,7,8-Tetrahydroxy[1]benzopyrano-[5,4,3-cde][1]benzopyran-5,10-dione; [1,2'-dicarboxylic acid, 4,4',5,5',6,6'-hexahydroxy-, di-.delta.-lactone; (1,1'-Biphenyl)-2,2'-dicarboxylic acid, 4,4',5,5',6,6'-hexahydroxy-, di-.delta.-lactone; 122328-15-8; 6,7,13,14-tetrahydroxy-2,9-dioxatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(14),4(16),5,7,11(15),12-hexaene-3,10-dione; 6,7,13,14-tetrahydroxy-2,9-dioxatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),4(16),5,7,11,13-hexaene-3,10-dione; 6,7,13,14-tetrahydroxy-2,9-dioxatetracyclo[6.6.2.0^{4,16}.0^{11,15}]hexadeca-1(15),4,6,8(16),11,13-hexaene-3,10-dione
|
|
| CAS | 476-66-4 | |
| PubChem CID | 5281855 | |
| ChEMBL ID | CHEMBL6246 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 302.19 | ALogp: | 1.1 |
| HBD: | 4 | HBA: | 8 |
| Rotatable Bonds: | 0 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 134.0 | Aromatic Rings: | 4 |
| Heavy Atoms: | 22 | QED Weighted: | 0.219 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -5.312 | MDCK Permeability: | 0.00001110 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0.006 |
| Human Intestinal Absorption (HIA): | 0.198 | 20% Bioavailability (F20%): | 0.073 |
| 30% Bioavailability (F30%): | 0.981 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.011 | Plasma Protein Binding (PPB): | 78.23% |
| Volume Distribution (VD): | 0.83 | Fu: | 23.96% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.788 | CYP1A2-substrate: | 0.088 |
| CYP2C19-inhibitor: | 0.013 | CYP2C19-substrate: | 0.043 |
| CYP2C9-inhibitor: | 0.233 | CYP2C9-substrate: | 0.098 |
| CYP2D6-inhibitor: | 0.006 | CYP2D6-substrate: | 0.123 |
| CYP3A4-inhibitor: | 0.053 | CYP3A4-substrate: | 0.011 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.346 | Half-life (T1/2): | 0.863 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0 | Human Hepatotoxicity (H-HT): | 0.144 |
| Drug-inuced Liver Injury (DILI): | 0.989 | AMES Toxicity: | 0.38 |
| Rat Oral Acute Toxicity: | 0.45 | Maximum Recommended Daily Dose: | 0.198 |
| Skin Sensitization: | 0.716 | Carcinogencity: | 0.314 |
| Eye Corrosion: | 0.009 | Eye Irritation: | 0.725 |
| Respiratory Toxicity: | 0.067 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
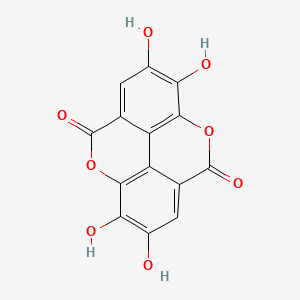
| ENC002516 | 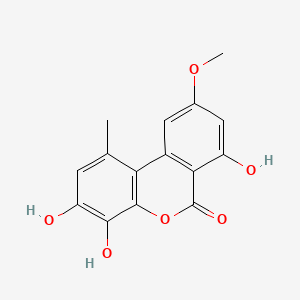 |
0.444 | D0K8KX | 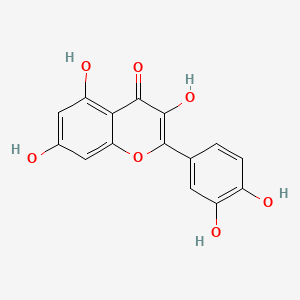 |
0.333 | ||
| ENC002018 | 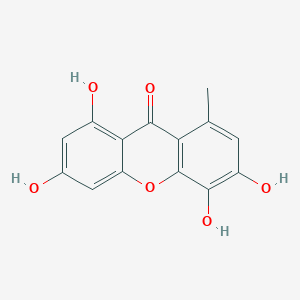 |
0.443 | D04AIT | 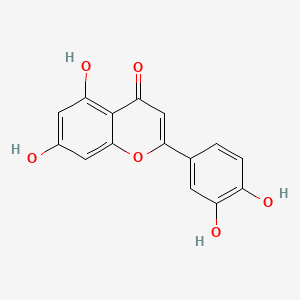 |
0.311 | ||
| ENC004844 | 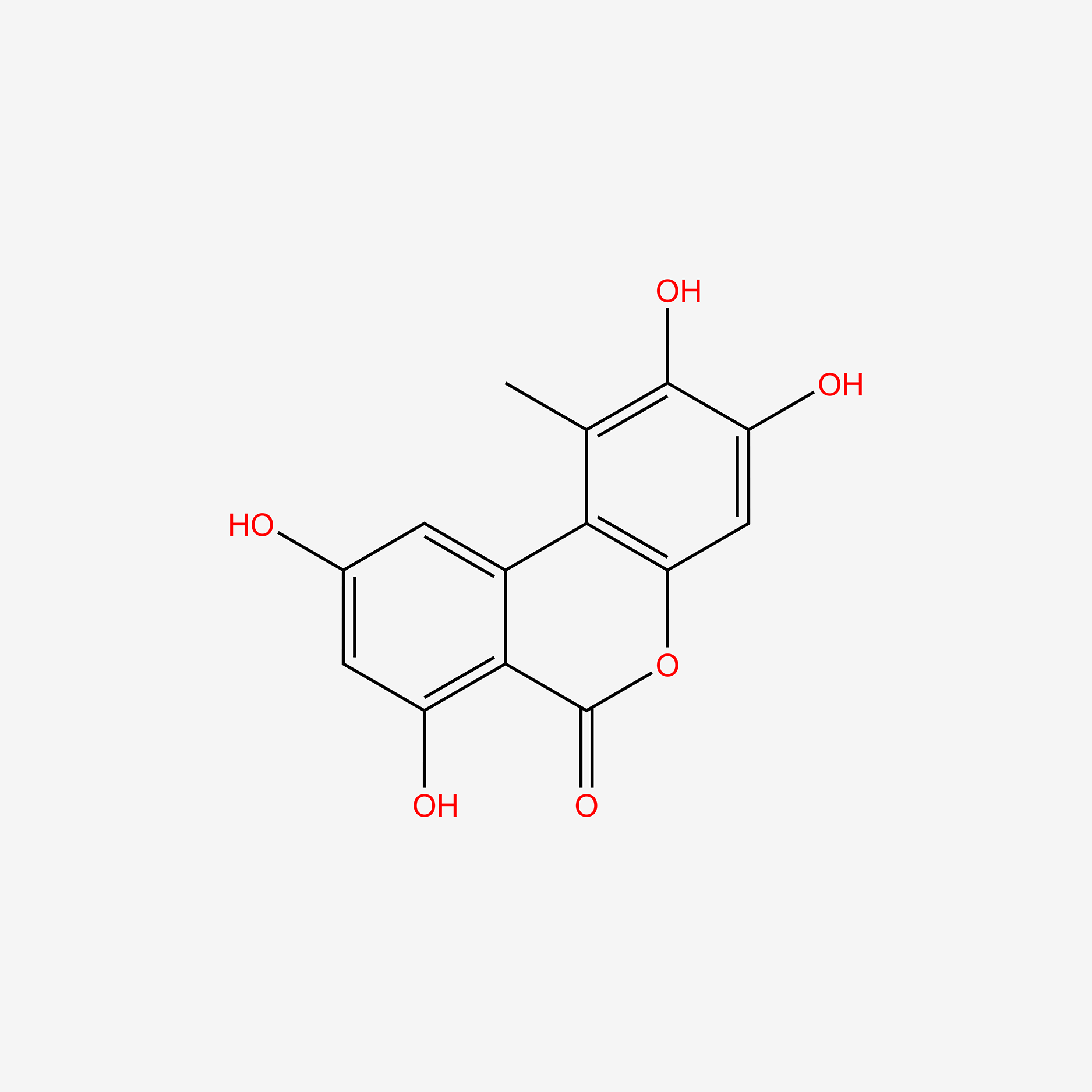 |
0.425 | D02TJS | 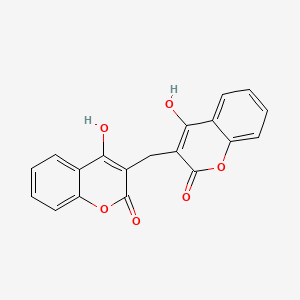 |
0.297 | ||
| ENC004389 | 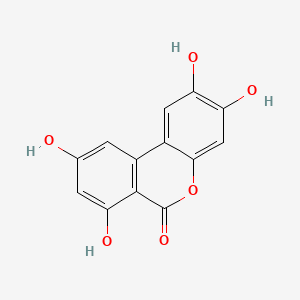 |
0.400 | D0FA2O |  |
0.244 | ||
| ENC001631 |  |
0.395 | D06GCK | 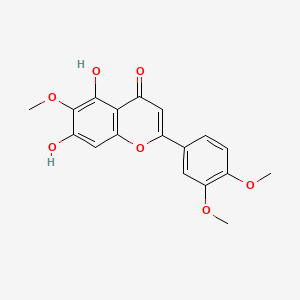 |
0.240 | ||
| ENC005647 |  |
0.393 | D07MGA |  |
0.235 | ||
| ENC004390 | 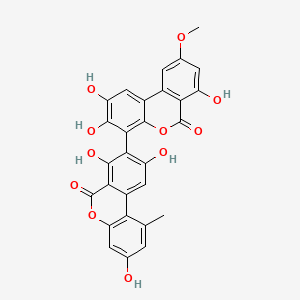 |
0.392 | D06FVX | 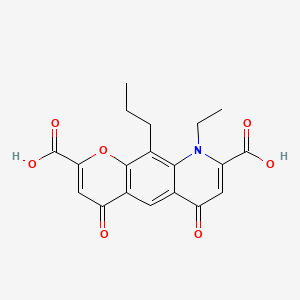 |
0.220 | ||
| ENC002024 | 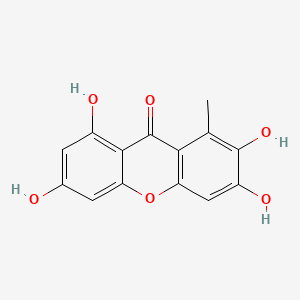 |
0.390 | D0AZ8C | 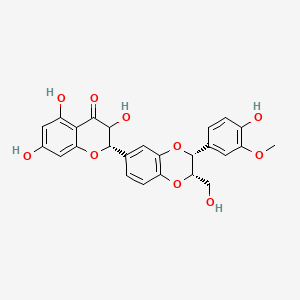 |
0.215 | ||
| ENC003471 | 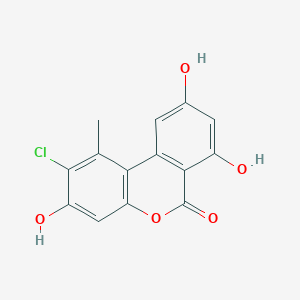 |
0.357 | D0G4KG | 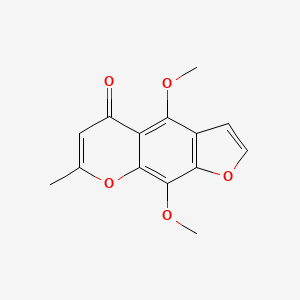 |
0.215 | ||
| ENC002811 | 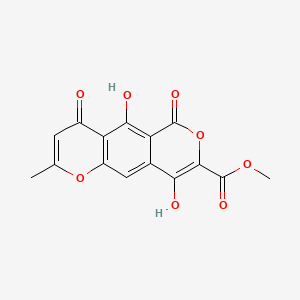 |
0.356 | D06NSS |  |
0.211 | ||