NPs Basic Information
|
Name |
Safranal
|
| Molecular Formula | C10H14O | |
| IUPAC Name* |
2,6,6-trimethylcyclohexa-1,3-diene-1-carbaldehyde
|
|
| SMILES |
CC1=C(C(CC=C1)(C)C)C=O
|
|
| InChI |
InChI=1S/C10H14O/c1-8-5-4-6-10(2,3)9(8)7-11/h4-5,7H,6H2,1-3H3
|
|
| InChIKey |
SGAWOGXMMPSZPB-UHFFFAOYSA-N
|
|
| Synonyms |
Safranal; 116-26-7; 2,3-Dihydro-2,2,6-trimethylbenzaldehyde; 1,3-CYCLOHEXADIENE-1-CARBOXALDEHYDE, 2,6,6-TRIMETHYL-; 2,6,6-trimethylcyclohexa-1,3-diene-1-carbaldehyde; 2,6,6-trimethylcyclohexa-1,3-dienecarbaldehyde; Dehydro-beta-cyclocitral; 2,6,6-trimethyl-1,3-cyclohexadiene-1-carboxaldehyde; (2,6,6-Trimethylcyclohexa-1,3-dienyl)methanal; FEMA No. 3389; 2,6,6-Trimethyl-1,3-cyclohexadienal; 2,6,6-Trimethylcyclohexa-1,3-dienyl methanal; 1,1,3-Trimethyl-2-formylcyclohexa-2,4-diene; CHEBI:53169; 4393FR07EA; EINECS 204-133-7; UNII-4393FR07EA; Dehydro-b-cyclocitral; SAFRANAL [MI]; Safranal (>80per cent); 2,6,6-Trimethylcyclohexa-1,3-dienylmethanal; DSSTox_CID_29357; DSSTox_RID_83473; DSSTox_GSID_49398; SCHEMBL23561; 1,3-Cyclohexadiene-1-carboxaldehyde,2,6,6-trimethyl-; Safranal, >=90%, stabilized; CHEMBL3183495; DTXSID7049398; FEMA 3389; SGAWOGXMMPSZPB-UHFFFAOYSA-; DEHYDRO-.BETA.-CYCLOCITRAL; HY-N7560; ZINC1851022; Tox21_202723; MFCD00209531; AKOS022504707; NCGC00260271-01; AS-75889; CAS-116-26-7; DB-019750; CS-0133689; FT-0631664; 1-Formyl-2,6,6-trimethyl-1,3-cyclohexadiene; C17062; D78038; EN300-781042; A803586; Q424919; 2,6,6-Trimethyl-1,3-cyclohexadiene-1-carbaldehyde; J-003414; 2,6,6-Trimethyl-1,3-cyclohexadiene-1-carbaldehyde #; 2,6,6-Trimethyl-1,3-cyclohexadienecarboxaldehyde, 9CI; Z1255446334; 2,6,6-TRIMETHYLCYCLOHEXA-1,3-DIENYL METHANAL [FHFI]
|
|
| CAS | 116-26-7 | |
| PubChem CID | 61041 | |
| ChEMBL ID | CHEMBL3183495 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 150.22 | ALogp: | 2.1 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 17.1 | Aromatic Rings: | 1 |
| Heavy Atoms: | 11 | QED Weighted: | 0.524 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.426 | MDCK Permeability: | 0.00003570 |
| Pgp-inhibitor: | 0.002 | Pgp-substrate: | 0.002 |
| Human Intestinal Absorption (HIA): | 0.006 | 20% Bioavailability (F20%): | 0.003 |
| 30% Bioavailability (F30%): | 0.001 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.403 | Plasma Protein Binding (PPB): | 82.27% |
| Volume Distribution (VD): | 2.228 | Fu: | 19.78% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.427 | CYP1A2-substrate: | 0.795 |
| CYP2C19-inhibitor: | 0.32 | CYP2C19-substrate: | 0.884 |
| CYP2C9-inhibitor: | 0.032 | CYP2C9-substrate: | 0.897 |
| CYP2D6-inhibitor: | 0.151 | CYP2D6-substrate: | 0.883 |
| CYP3A4-inhibitor: | 0.032 | CYP3A4-substrate: | 0.298 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 2.445 | Half-life (T1/2): | 0.6 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.019 | Human Hepatotoxicity (H-HT): | 0.276 |
| Drug-inuced Liver Injury (DILI): | 0.099 | AMES Toxicity: | 0.301 |
| Rat Oral Acute Toxicity: | 0.594 | Maximum Recommended Daily Dose: | 0.513 |
| Skin Sensitization: | 0.861 | Carcinogencity: | 0.889 |
| Eye Corrosion: | 0.937 | Eye Irritation: | 0.971 |
| Respiratory Toxicity: | 0.965 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
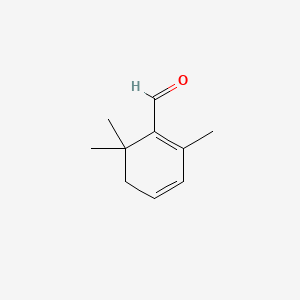
| ENC000328 | 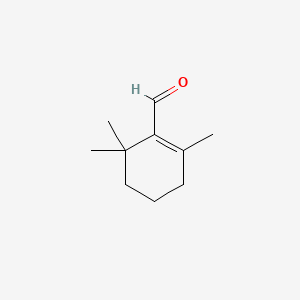 |
0.415 | D0K7LU | 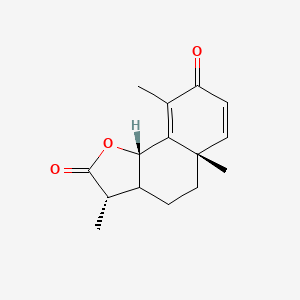 |
0.200 | ||
| ENC001457 | 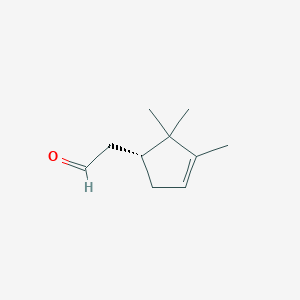 |
0.289 | D0U4VT |  |
0.200 | ||
| ENC000146 | 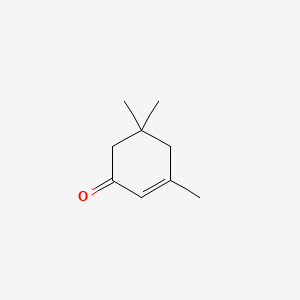 |
0.279 | D0G3PI | 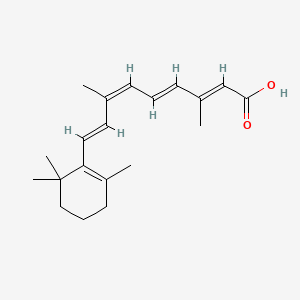 |
0.189 | ||
| ENC001425 | 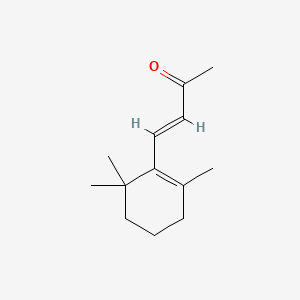 |
0.269 | D02DGU | 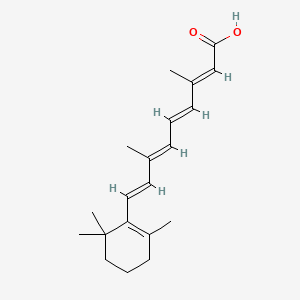 |
0.189 | ||
| ENC001898 | 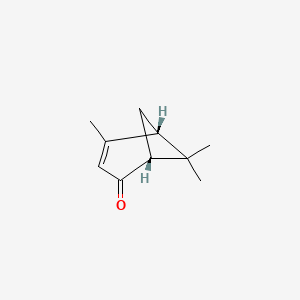 |
0.261 | D00DKK | 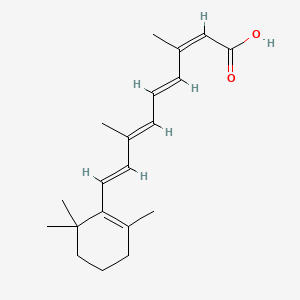 |
0.189 | ||
| ENC005031 |  |
0.261 | D0H1QY | 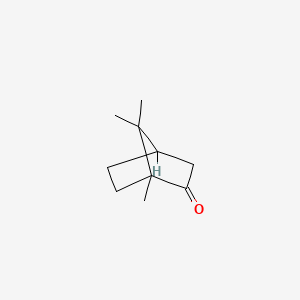 |
0.184 | ||
| ENC000552 |  |
0.244 | D0H6VY | 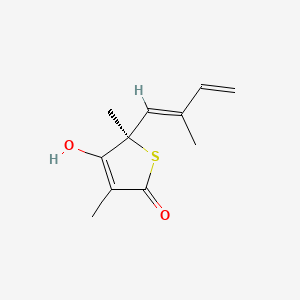 |
0.182 | ||
| ENC000649 |  |
0.244 | D0E9CD | 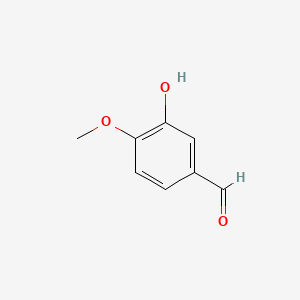 |
0.180 | ||
| ENC001334 |  |
0.244 | D0S7WX | 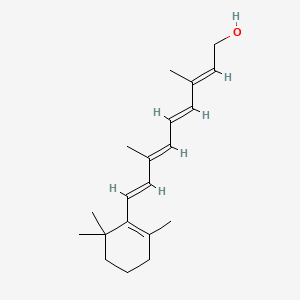 |
0.178 | ||
| ENC000412 | 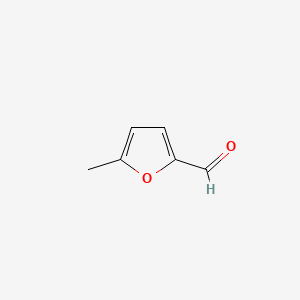 |
0.244 | D0Q4XQ |  |
0.170 | ||