NPs Basic Information

|
Name |
1,4-Cineole
|
| Molecular Formula | C10H18O | |
| IUPAC Name* |
1-methyl-4-propan-2-yl-7-oxabicyclo[2.2.1]heptane
|
|
| SMILES |
CC(C)C12CCC(O1)(CC2)C
|
|
| InChI |
InChI=1S/C10H18O/c1-8(2)10-6-4-9(3,11-10)5-7-10/h8H,4-7H2,1-3H3
|
|
| InChIKey |
RFFOTVCVTJUTAD-UHFFFAOYSA-N
|
|
| Synonyms |
1,4-Cineole; 470-67-7; Isocineole; 1,4-Cineol; 1,4-EPOXY-P-MENTHANE; p-Menthane, 1,4-epoxy-; FEMA No. 3658; Isocineple; p-Menthane, 1,4-epoxy; 1-methyl-4-propan-2-yl-7-oxabicyclo[2.2.1]heptane; 1-Methyl-4-(1-methylethyl)-7-oxabicyclo(2.2.1)heptane; 1-Isopropyl-4-methyl-7-oxabicyclo(2.2.1)heptane; 1-Isopropyl-4-methyl-7-oxabicyclo[2.2.1]heptane; 7-Oxabicyclo(2.2.1)heptane, 1-isopropyl-4-methyl-; 1-methyl-4-(propan-2-yl)-7-oxabicyclo[2.2.1]heptane; B55JTU839B; 7-Oxabicyclo(2.2.1)heptane, 1-methyl-4-(1-methylethyl)-; CHEBI:80788; 1-METHYL-4-(1-METHYLETHYL)-7-OXABICYCLO[2.2.1]HEPTANE; 7-Oxabicyclo[2.2.1]heptane, 1-methyl-4-(1-methylethyl)-; 4-isopropyl-1-methyl-7-oxabicyclo[2.2.1]heptane; 1,4-Cineole (natural); HSDB 5425; EINECS 207-428-9; BRN 0104974; UNII-B55JTU839B; IsocineoleIsocineole; 5-17-01-00273 (Beilstein Handbook Reference); 1beta,4beta-Epoxy-p-menthane; SCHEMBL231925; 1,4-CINEOLE [FHFI]; 1,4-Cineole, >=85%; CHEMBL2288022; DTXSID3047396; SCHEMBL13180469; SCHEMBL15450554; SCHEMBL18993155; 1,4-Cineole, analytical standard; 7-Oxabicyclo(2.2.1)heptane, 1-isopropyl-4-methyl- (6CI); 1,4-Cineole, >=95%, FG; HY-N7117; ZINC2040513; (+/-)-1,4-CINEOLE; MFCD00209502; s5419; 1,4-EPOXY-P-MENTHANE [HSDB]; CCG-266250; AS-81210; 1-isopropyl-4-methylbicyclo[2.2.1]heptane; C3652; CS-0066997; C16909; 1-Isopropyl-4-methyl-7-oxabicyclo[2.2.1]heptane #; 4-methyl-1-propan-2-yl-7-oxabicyclo[2.2.1]heptane; Q12470088; 1-Methyl-4-(1-methylethyl)-7-oxabicyclo[2.2.1]heptane, 9CI; (1S,4S)-1-ISOPROPYL-4-METHYL-7-OXABICYCLO[2.2.1]HEPTANE; (1s,4s)-1-methyl-4-(propan-2-yl)-7-oxabicyclo[2.2.1]heptane
|
|
| CAS | 470-67-7 | |
| PubChem CID | 10106 | |
| ChEMBL ID | CHEMBL2288022 |
*Note: the IUPAC Name was collected from PubChem.
Chemical Classification: |
|
|
|---|
——————————————————————————————————————————
NPs Species Source
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference | |
|---|---|---|---|---|---|---|---|---|
| Endophyte ID | Endophyte Name | Family | Genus | Taxonomy ID | GenBank ID | Closest GenBank ID | Reference |
NPs Biological Activity
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bioactivity Name | Target ID | Target Name | Target Type | Target Organism | Target Organism ID | Potency of Bioactivity | Activity Type | Value | Unit | Endophyte ID | Endophyte Name |
NPs Physi-Chem Properties
| Molecular Weight: | 154.25 | ALogp: | 2.5 |
| HBD: | 0 | HBA: | 1 |
| Rotatable Bonds: | 1 | Lipinski's rule of five: | Accepted |
| Polar Surface Area: | 9.2 | Aromatic Rings: | 2 |
| Heavy Atoms: | 11 | QED Weighted: | 0.561 |
——————————————————————————————————————————
NPs ADMET Properties*
ADMET: Absorption
| Caco-2 Permeability: | -4.355 | MDCK Permeability: | 0.00001600 |
| Pgp-inhibitor: | 0 | Pgp-substrate: | 0 |
| Human Intestinal Absorption (HIA): | 0.003 | 20% Bioavailability (F20%): | 0.009 |
| 30% Bioavailability (F30%): | 0.008 |
——————————————————————————————————————————
ADMET: Distribution
| Blood-Brain-Barrier Penetration (BBB): | 0.886 | Plasma Protein Binding (PPB): | 90.09% |
| Volume Distribution (VD): | 2.34 | Fu: | 7.14% |
——————————————————————————————————————————
ADMET: Metabolism
| CYP1A2-inhibitor: | 0.094 | CYP1A2-substrate: | 0.696 |
| CYP2C19-inhibitor: | 0.419 | CYP2C19-substrate: | 0.933 |
| CYP2C9-inhibitor: | 0.208 | CYP2C9-substrate: | 0.867 |
| CYP2D6-inhibitor: | 0.022 | CYP2D6-substrate: | 0.571 |
| CYP3A4-inhibitor: | 0.022 | CYP3A4-substrate: | 0.217 |
——————————————————————————————————————————
ADMET: Excretion
| Clearance (CL): | 9.344 | Half-life (T1/2): | 0.371 |
——————————————————————————————————————————
ADMET: Toxicity
| hERG Blockers: | 0.026 | Human Hepatotoxicity (H-HT): | 0.384 |
| Drug-inuced Liver Injury (DILI): | 0.045 | AMES Toxicity: | 0.008 |
| Rat Oral Acute Toxicity: | 0.058 | Maximum Recommended Daily Dose: | 0.02 |
| Skin Sensitization: | 0.154 | Carcinogencity: | 0.187 |
| Eye Corrosion: | 0.241 | Eye Irritation: | 0.954 |
| Respiratory Toxicity: | 0.752 |
——————————————————————————————————————————
*Note: the ADMET properties was calculated by ADMETlab 2.0. Reference: PMID: 33893803.
Similar Compounds*
Compounds similar to EMNPD with top10 similarity:
| Similar NPs | Similar Drugs | ||||||
|---|---|---|---|---|---|---|---|
| NPs ID | NPs 2D Structure | Similarity Score | TTD ID | Drug 2D Structure | Similarity Score | ||
| ENC000085 | 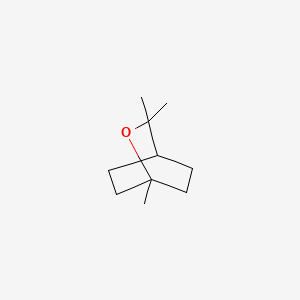 |
0.364 | D07QKN | 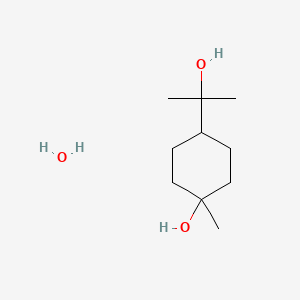 |
0.216 | ||
| ENC005519 | 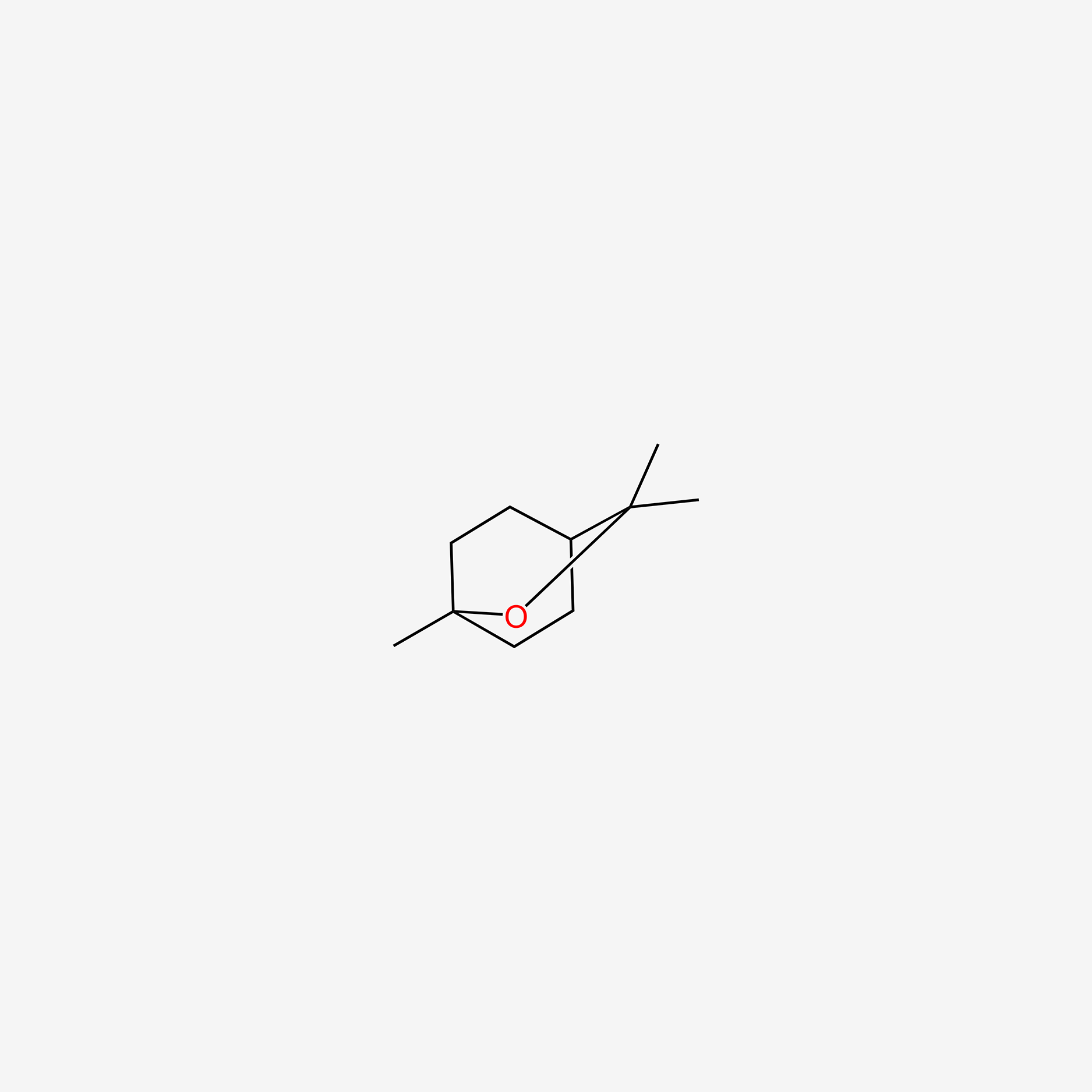 |
0.364 | D0H1QY | 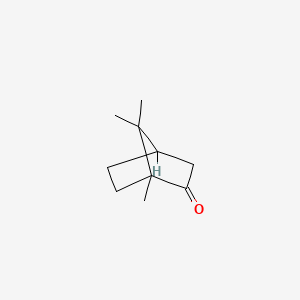 |
0.204 | ||
| ENC002232 |  |
0.341 | D04CSZ |  |
0.180 | ||
| ENC000653 | 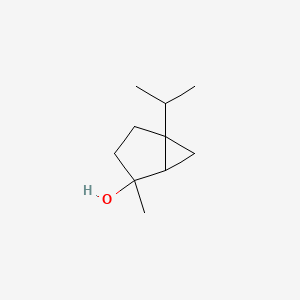 |
0.341 | D01CKY | 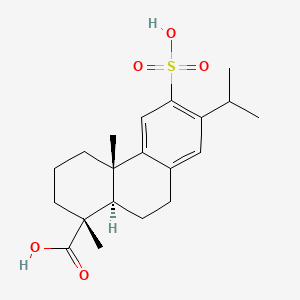 |
0.179 | ||
| ENC005252 |  |
0.313 | D08KVZ |  |
0.174 | ||
| ENC000360 |  |
0.313 | D0V8HA | 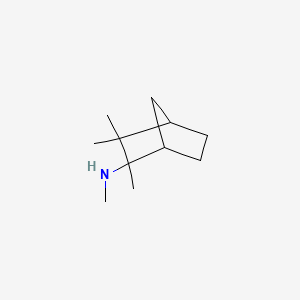 |
0.170 | ||
| ENC003502 | 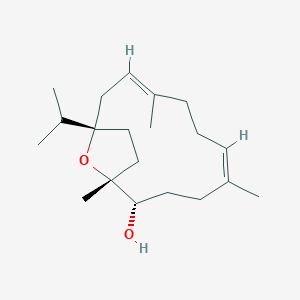 |
0.304 | D0Z1XD | 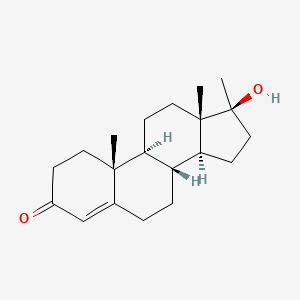 |
0.167 | ||
| ENC001077 |  |
0.259 | D0U3GL | 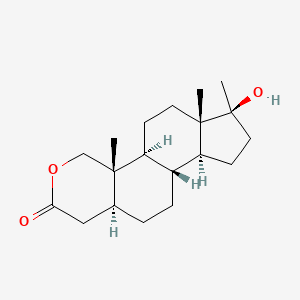 |
0.167 | ||
| ENC002415 |  |
0.258 | D04ATM |  |
0.165 | ||
| ENC001341 | 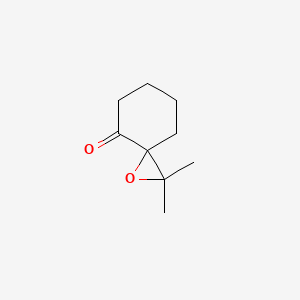 |
0.255 | D0S3WH |  |
0.162 | ||